 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aco004459.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Bromeliaceae; Ananas
|
||||||||
| Family | Nin-like | ||||||||
| Protein Properties | Length: 992aa MW: 109147 Da PI: 6.5902 | ||||||||
| Description | Nin-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | RWP-RK | 90.7 | 1.1e-28 | 641 | 692 | 1 | 52 |
RWP-RK 1 aekeisledlskyFslpikdAAkeLgvclTvLKriCRqyGIkRWPhRkiksl 52
aek+isle+l++yF++++k+AAk+Lgvc+T++KriCRq+GI RWP+Rki+++
Aco004459.1 641 AEKTISLEVLQQYFAGSLKNAAKSLGVCPTTMKRICRQHGILRWPSRKINKV 692
589***********************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51519 | 17.086 | 631 | 712 | IPR003035 | RWP-RK domain |
| Pfam | PF02042 | 4.6E-25 | 644 | 692 | IPR003035 | RWP-RK domain |
| SuperFamily | SSF54277 | 7.85E-22 | 885 | 980 | No hit | No description |
| SMART | SM00666 | 4.2E-21 | 896 | 978 | IPR000270 | PB1 domain |
| PROSITE profile | PS51745 | 20.981 | 896 | 978 | IPR000270 | PB1 domain |
| Pfam | PF00564 | 1.4E-15 | 897 | 977 | IPR000270 | PB1 domain |
| Gene3D | G3DSA:3.10.20.240 | 5.0E-22 | 897 | 977 | No hit | No description |
| CDD | cd06407 | 4.29E-33 | 897 | 977 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0010118 | Biological Process | stomatal movement | ||||
| GO:0010167 | Biological Process | response to nitrate | ||||
| GO:0042128 | Biological Process | nitrate assimilation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 992 aa Download sequence Send to blast |
MESPPSPRPL PPPPPPPPLP RWCFLLRSMP GRDEEAAEAR GALMDLDDLD LDLDLDLDPD 60 PPSCLLPPRR PAPAADDAEW PFDALVSPSL LSSSSPPQSL LHLSFPSPPS PLCLFEDRPF 120 DASTSIAELA LYNPDTVSRN INGTNDKRRP LQLLVEEDNA DHTYIIKERM TQALRQFKES 180 TDQHLLVQVW APVKNGDRYV LTTSGQPFVL DHQSVALLQY RTVSLTYMFS VDGDNDGELG 240 LPGRVYRQKV PEWTPNVQYY SSKEYPRLCH ALNYNVRGTV ALPVFYPSAH TCVAVIELIM 300 TSQKINYAGE VGKVCKALEA VNLKSTEIQD HPSVQAKHPL SHVFVFLQGR FFLPNQICNK 360 GRQTALAEIL ETLAVVCEDH KLPLAQTWVP CRHRSVLAHG GGLRKSCSSF DGSCMGQVCM 420 STSDVAFHVI DAHIWRFRDA CVEHHLQKGQ GVAGIAFESR RPCFSEDITQ FSKTQYPLVH 480 YSRMCGLAGC FAVCLQSTFT GNDDYILEFF LPPECKNAGE QRVLLEAMII SMKRCFRSLK 540 VVADVELQEG ITLQSIDMLV IDNREFASGH PRASLECNFD KGFNGAGKLD NRVSDMPEKH 600 ILANGNAVNN GIVIEQNGSG TSSSSLANKN NKPSERRRGK AEKTISLEVL QQYFAGSLKN 660 AAKSLGVCPT TMKRICRQHG ILRWPSRKIN KVNRSLSKLK QVIESVDGAF TLPSLPCPLP 720 VAVGSLPSPS NVDKTNKTKE SEPLKPFTLH TERENKLSTR DTSPDNDGSG KKLFPHCSRS 780 GSSSEEGSSN THTSEGSCHA SPQNENVVSG PFNSTFLGPE LKPSCTLGLI NPNTNFICLN 840 TTAVEPQSPL NGMLVEDSGS SKYLKNLCPC AVEIPHDEHN VGPTSNNPTS MANSRVVTIK 900 ASYKDDIVRF RFTVAGNVTE LKDEIAKRLK LEVGMFDVKY LDDDPEWVLL ACNADLEECV 960 EIARLSGRHV VRLLVSDISA SLGSSCESSR G* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00449 | DAP | Transfer from AT4G24020 | Download |
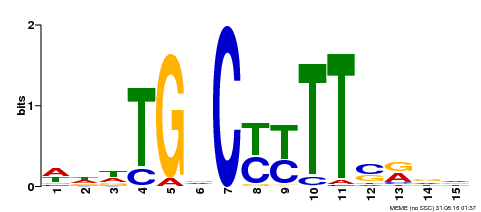
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020088915.1 | 0.0 | protein NLP3-like | ||||
| Swissprot | Q5NB82 | 0.0 | NLP3_ORYSJ; Protein NLP3 | ||||
| TrEMBL | A0A199W4Y8 | 0.0 | A0A199W4Y8_ANACO; Protein NLP3 | ||||
| STRING | OPUNC01G08520.1 | 0.0 | (Oryza punctata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP4778 | 36 | 60 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G24020.1 | 0.0 | NIN like protein 7 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aco004459.1 |



