 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aco006038.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Bromeliaceae; Ananas
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 265aa MW: 29210.1 Da PI: 4.8915 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 92.1 | 4.4e-29 | 68 | 126 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-ST.T---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsa.gCpvkkkversaedpkvveitYegeHnhe 59
+D ++WrKYGqK++kgs++pr+YYrC+s+ gCp++k+vers+ dp+++++tY+ +Hnh+
Aco006038.1 68 SDPWAWRKYGQKPIKGSPYPRGYYRCSSSkGCPARKQVERSRVDPTTLVVTYSFDHNHP 126
699************************998****************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50811 | 31.617 | 62 | 128 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 1.8E-30 | 63 | 128 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 2.35E-25 | 64 | 128 | IPR003657 | WRKY domain |
| SMART | SM00774 | 8.8E-34 | 67 | 127 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 3.5E-25 | 69 | 126 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 265 aa Download sequence Send to blast |
MDGYYYSSDS PDDAAVPSPP GEPKPTSHPP PPPTKRSRRS VQKRVVSVPI AEADPSRGKG 60 AGESGPPSDP WAWRKYGQKP IKGSPYPRGY YRCSSSKGCP ARKQVERSRV DPTTLVVTYS 120 FDHNHPWPAP KNPHRPSSKR PARAGEEREP EAGRAGPDEP EEEEEEEVGA RGRFGDLMIG 180 AEFEWRAEEE DEAAVLLYGP MYGAALLPEE RGDSGRRAEE EDALFAGLGE LPECAVVFRH 240 RRGFGPEPEP EPEPEPEPWR SMNS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5w3x_B | 8e-19 | 43 | 129 | 6 | 80 | Disease resistance protein RRS1 |
| 5w3x_D | 8e-19 | 43 | 129 | 6 | 80 | Disease resistance protein RRS1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00167 | DAP | Transfer from AT1G29280 | Download |
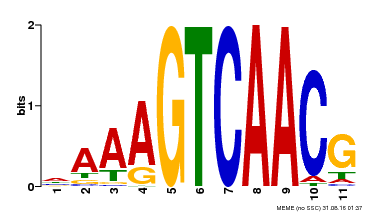
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020105327.1 | 0.0 | probable WRKY transcription factor 65 | ||||
| Swissprot | Q9LP56 | 1e-57 | WRK65_ARATH; Probable WRKY transcription factor 65 | ||||
| TrEMBL | A0A199UDG9 | 1e-141 | A0A199UDG9_ANACO; Putative WRKY transcription factor 65 | ||||
| STRING | Gorai.011G037500.1 | 9e-66 | (Gossypium raimondii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP7088 | 36 | 51 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G58710.2 | 7e-40 | WRKY DNA-binding protein 69 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aco006038.1 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



