 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aco006221.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Bromeliaceae; Ananas
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 337aa MW: 36881.8 Da PI: 6.6147 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 106.3 | 1.6e-33 | 206 | 263 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
+Dgy+WrKYGqK vk+s++prsYYrCt+++C+vkk+vers+edp++v++tYeg+H+h+
Aco006221.1 206 EDGYRWRKYGQKAVKNSPYPRSYYRCTTQKCSVKKRVERSHEDPSIVITTYEGRHTHQ 263
8********************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 2.2E-35 | 190 | 265 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.31E-29 | 197 | 265 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 31.617 | 200 | 265 | IPR003657 | WRKY domain |
| SMART | SM00774 | 8.4E-39 | 205 | 264 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.3E-26 | 206 | 263 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 337 aa Download sequence Send to blast |
MSQERGELYH HQHGHLFLNE MINSGGDLSC FFSHHEAMSV ADAAPFAGFA DYDYGAALSR 60 AAFGDVRCDE LGLFDAPPAK PELLIDVDAA IDYGAVVGRR HGLDVSIGGG GGGGDGGTGP 120 VTPNSSVSSS SSEAGGDEES GGRSKKDRIK EEEEEGGEEK QLQARGSDDQ GGEKSKKLVK 180 AKNKGEKRQR EPRFAFMTKS EVDHLEDGYR WRKYGQKAVK NSPYPRSYYR CTTQKCSVKK 240 RVERSHEDPS IVITTYEGRH THQSPANLRG SSHFLAPSPS FRAQEFLNPS SHHHMSTNPT 300 MYLPISLPPP LQQLQVLPDY GLLQDIIPSF IHQNQP* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 8e-28 | 196 | 266 | 8 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 8e-28 | 196 | 266 | 8 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00440 | DAP | Transfer from AT4G18170 | Download |
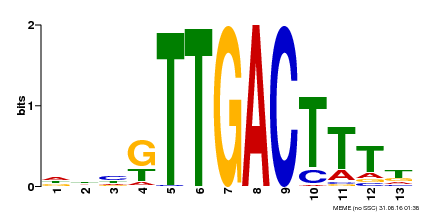
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020105446.1 | 0.0 | probable WRKY transcription factor 71 | ||||
| TrEMBL | A0A199VFX6 | 0.0 | A0A199VFX6_ANACO; Putative WRKY transcription factor 28 | ||||
| STRING | XP_008798309.1 | 1e-101 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP623 | 37 | 173 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G18170.1 | 2e-50 | WRKY DNA-binding protein 28 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aco006221.1 |



