 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aco007331.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Bromeliaceae; Ananas
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 996aa MW: 109537 Da PI: 6.4395 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 129.5 | 1.2e-40 | 147 | 224 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+CqvegC +dls+ k+yhrrhkvCe+h+ka++++v + qrfCqqCsrfh+l+efDe+krsCrrrLa+hn+rrrk+++
Aco007331.1 147 VCQVEGCGTDLSTSKDYHRRHKVCEMHAKATTAVVGNAVQRFCQQCSRFHHLQEFDEGKRSCRRRLAGHNRRRRKTHP 224
6**************************************************************************976 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 1.4E-32 | 142 | 209 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.146 | 145 | 222 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 7.46E-38 | 146 | 226 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 4.6E-30 | 148 | 221 | IPR004333 | Transcription factor, SBP-box |
| CDD | cd00204 | 4.02E-9 | 748 | 879 | No hit | No description |
| SuperFamily | SSF48403 | 1.27E-8 | 752 | 880 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 5.0E-7 | 774 | 878 | IPR020683 | Ankyrin repeat-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 996 aa Download sequence Send to blast |
MEAQLGGGSN HFYGNGVLER NGIGKRNFEW DLNDWKWDGD LFLASPLNGG LTDRTNKQLI 60 TNAANGLPSN SSSSGSEETD LVIVRPGYAE SEKRKRTVDL EEAEPSDGNG SLSLKLGGHE 120 YPTVEANGED KNGKKSKSAG CSTNRPVCQV EGCGTDLSTS KDYHRRHKVC EMHAKATTAV 180 VGNAVQRFCQ QCSRFHHLQE FDEGKRSCRR RLAGHNRRRR KTHPDAAVGG TTSDEKSSSS 240 LLMSLLKILT NLHSDSAEPA NNQDLVSHFL RNLANLAGLL DATKLAGLLQ SSQSLQKLGN 300 SAGTSSDVAN ALILNSASAP EASRKLVCSA SAVACVSAAQ DPCKLSGASV PCAIPTECTP 360 ARADTIRTEP TVHKARLKDF DLNSTYGDTQ ECGEAYEQSV NPAYIVNGSP TGPSWLLQDS 420 RQLSPPQTSG TSYSTSEKSQ SSSNGDGQCR TDRIVFKLFG KDPNDLPLVL RAQILDWLSN 480 SPTDIESYIR PGCIILTVYL RLTNSALEEL CNSLSSYLER LLNSSANDFW RSGWIYARIE 540 DQIAFIHNGQ ILLEAPLPLA YHDHCEIACV SPIAVPHSSK VTFTVKGFNL ARSATRILCS 600 FEGKCLVEET TKQSVLEETD EDTQQEGPEC LSFSCSLPES RGRGFIEVED ETLSNAFFPF 660 IVADQELCSE ICILESSIDV VDDCSEERSD TEIARKQALY FINELGWLLR KTNLRTKYQK 720 MEVYPALFHL RRFQWLISFA MEHDWCAVIK QLLDILFSGT VDLDGKSPRE IALSENLLHT 780 AVRRNCKAMV KVLLKYTPPV KNSEDKFEKL LFRPDVVGPS NLTPLHIAAA TSGVEDVLDA 840 LTDDPDMRGI TAWKSARDNN GFTPEDYARV QGHESYLHLV RKKTDRELDK GHVVIGIPGN 900 LCAKFANDPR PVNSSFEISK NKLASSAPAP YCNRCSMQMA HRSLGTRTLL YRPLILSMVG 960 IAAVCVCVGL LFKSPPTVLY VFPSFRWELL TYGSM* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 3e-29 | 142 | 221 | 5 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00633 | PBM | Transfer from PK06791.1 | Download |
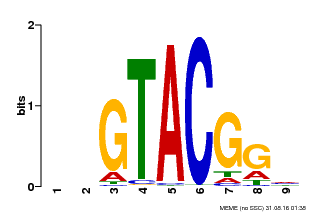
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020113769.1 | 0.0 | squamosa promoter-binding-like protein 6 | ||||
| Refseq | XP_020113770.1 | 0.0 | squamosa promoter-binding-like protein 6 | ||||
| Swissprot | Q75LH6 | 0.0 | SPL6_ORYSJ; Squamosa promoter-binding-like protein 6 | ||||
| TrEMBL | A0A199V0C1 | 0.0 | A0A199V0C1_ANACO; Squamosa promoter-binding-like protein 6 | ||||
| STRING | XP_008813721.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1998 | 38 | 92 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G47070.1 | 0.0 | squamosa promoter binding protein-like 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aco007331.1 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



