 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aco009397.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Bromeliaceae; Ananas
|
||||||||
| Family | SRS | ||||||||
| Protein Properties | Length: 335aa MW: 34947.1 Da PI: 8.4123 | ||||||||
| Description | SRS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF702 | 210.8 | 3.4e-65 | 100 | 250 | 5 | 153 |
DUF702 5 tasCqdCGnqakkdCaheRCRtCCksrgfdCathvkstWvpaakrrerqqqlaaasskaaasa..aeaaskrkrelkskkqsalsstklssaeskkel 100
++sCqdCGnqakkdCah+RCRtCCksrgf+CathvkstWvpaa+rr+rq+q++ + + +s+ a+ +++r+r+ +s+ ++s +
Aco009397.1 100 GVSCQDCGNQAKKDCAHQRCRTCCKSRGFHCATHVKSTWVPAARRRDRQHQHQLQLAALPSSSspADPSKRRPRDRRSSLPPPSTSGGGGGGGLGAGA 197
679*********************************************88777665555544422444555556554444444444444445555666 PP
DUF702 101 etsslPeevsseavfrcvrvssvddgeeelaYqtavsigGhvfkGiLydqGle 153
+++P+evsseavfrcvr++ vd++e+e+aYq++vsigGhvfkGiL+dqG++
Aco009397.1 198 VEARFPAEVSSEAVFRCVRIGPVDEAEDEYAYQATVSIGGHVFKGILRDQGPD 250
6777***********************************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05142 | 4.5E-62 | 100 | 250 | IPR007818 | Protein of unknown function DUF702 |
| TIGRFAMs | TIGR01623 | 3.0E-27 | 102 | 144 | IPR006510 | Zinc finger, lateral root primordium type 1 |
| TIGRFAMs | TIGR01624 | 5.4E-25 | 201 | 249 | IPR006511 | Lateral Root Primordium type 1, C-terminal |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010051 | Biological Process | xylem and phloem pattern formation | ||||
| GO:0010252 | Biological Process | auxin homeostasis | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0048479 | Biological Process | style development | ||||
| GO:0048480 | Biological Process | stigma development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0046982 | Molecular Function | protein heterodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 335 aa Download sequence Send to blast |
MAGFPLGGGG SSGHIPTSRD DQNPLPPDTF FLYARAGGTG TSGFQLWQQQ QQQQQQQQQQ 60 QQQQAQFYSS SGAILSFSHE ASASGSGQSG GRYGGGGGSG VSCQDCGNQA KKDCAHQRCR 120 TCCKSRGFHC ATHVKSTWVP AARRRDRQHQ HQLQLAALPS SSSPADPSKR RPRDRRSSLP 180 PPSTSGGGGG GGLGAGAVEA RFPAEVSSEA VFRCVRIGPV DEAEDEYAYQ ATVSIGGHVF 240 KGILRDQGPD LSSYSYSGYH YHTQPPQPGE SSSSAAAAAA AAAASAEAAI TSASAAAAPA 300 AVMDPYPTPL SAFAAGTQFF PQQHHHHHHH HPRS* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 89 | 97 | GGRYGGGGG |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00613 | PBM | Transfer from AT3G51060 | Download |
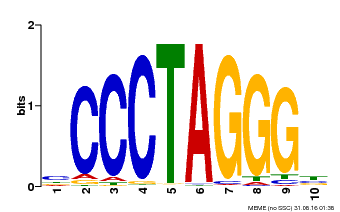
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020113236.1 | 0.0 | protein SHI RELATED SEQUENCE 1-like isoform X1 | ||||
| TrEMBL | A0A199VLD0 | 0.0 | A0A199VLD0_ANACO; Protein SHI RELATED SEQUENCE 1 | ||||
| STRING | GSMUA_Achr4P17960_001 | 2e-72 | (Musa acuminata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1354 | 35 | 124 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G75520.1 | 2e-35 | SHI-related sequence 5 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aco009397.1 |



