 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aco009781.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Bromeliaceae; Ananas
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 459aa MW: 48741.5 Da PI: 5.5099 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 101.4 | 5.9e-32 | 242 | 297 | 1 | 56 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRla 56
k+r++W+p+LH+rFv+a++ LGGs++AtPk+i+elmkv+gLt+++vkSHLQkYRl+
Aco009781.1 242 KARRCWSPDLHRRFVNALQMLGGSQVATPKQIRELMKVDGLTNDEVKSHLQKYRLH 297
68****************************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 13.355 | 239 | 299 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 8.96E-17 | 239 | 300 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 3.1E-28 | 240 | 300 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.3E-26 | 242 | 297 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 8.0E-7 | 244 | 295 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009266 | Biological Process | response to temperature stimulus | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010452 | Biological Process | histone H3-K36 methylation | ||||
| GO:0048579 | Biological Process | negative regulation of long-day photoperiodism, flowering | ||||
| GO:1903507 | Biological Process | negative regulation of nucleic acid-templated transcription | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 459 aa Download sequence Send to blast |
MASPSDLSLD YKPNNYPIIQ KPVGEHPNQT QKIQEFLARL EEERLKIEAF KRELPLCLQL 60 LNNAVEAYRQ QLEAYQTSQG PRPVLEEFIP VKQMSVDGLE TPNGPSSEKA SWMISAQLWS 120 NPAESKPQQA PKGGGDVSPK LGLLDAKQRN GGGGAFLPFS KEKAKGGGGV TPRGLPELAL 180 ASADKGEEEK KGGVAVMENG GFGGRRENGG KGVGTEQAKG GNVAEGAAAA AAVAPPPQTH 240 RKARRCWSPD LHRRFVNALQ MLGGSQVATP KQIRELMKVD GLTNDEVKSH LQKYRLHTRR 300 PIPAPPASAA AAPQLVVLGG IWVPPEYATT AAGPAIYGAH PGPAHYCTAP VPQDFYPTAT 360 GVAPLPHHQI HPALHRSAAT ATVSAVPTAY RGHRGSPESD MRSGGEQSME SIEDEEEEEE 420 EEGGEQRAAE EEGDEGVAVE EAMKAGEGDG GDDVALKF* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4r_A | 3e-14 | 242 | 295 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 3e-14 | 242 | 295 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 3e-14 | 242 | 295 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 3e-14 | 242 | 295 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor acting as a flowering repressor, directly repressing FT expression in a dosage-dependent manner in the leaf vasculature. {ECO:0000269|PubMed:25132385}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00261 | DAP | Transfer from AT2G03500 | Download |
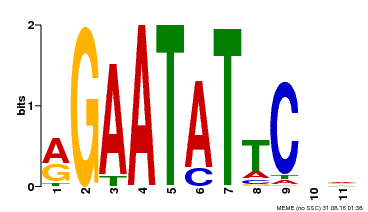
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by SVP. Down-regulated by high temperature and gibberellic acid treatment. Not regulated by photoperiod, circadian rhythm under long days or vernalization. {ECO:0000269|PubMed:25132385}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020097321.1 | 0.0 | myb family transcription factor EFM | ||||
| Swissprot | Q9ZQ85 | 1e-111 | EFM_ARATH; Myb family transcription factor EFM | ||||
| TrEMBL | A0A199VKQ5 | 0.0 | A0A199VKQ5_ANACO; Putative transcription factor GLK1 | ||||
| STRING | XP_008792133.1 | 1e-175 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP9773 | 33 | 38 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G03500.1 | 1e-102 | G2-like family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aco009781.1 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



