 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aco011363.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Bromeliaceae; Ananas
|
||||||||
| Family | LBD | ||||||||
| Protein Properties | Length: 270aa MW: 27157.6 Da PI: 8.4872 | ||||||||
| Description | LBD family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF260 | 140 | 7.6e-44 | 48 | 147 | 1 | 100 |
DUF260 1 aCaaCkvlrrkCakdCvlapyfpaeqpkkfanvhklFGasnvlkllkalpeeeredamsslvyeAearardPvyGavgvilklqqqleqlkaelallk 98
+CaaCk+lrrkC++dCv+apyfp ++p+kf vh++FGasnv+kll++l++ +reda++sl+yeA++r+rdPvyG+vgvi+ lq+ql+ql+ +l ++k
Aco011363.1 48 PCAACKFLRRKCQPDCVFAPYFPPDNPQKFVHVHRVFGASNVTKLLNELHPYQREDAVNSLAYEADMRLRDPVYGCVGVISILQHQLRQLQMDLTTAK 145
7*********************************************************************************************9999 PP
DUF260 99 ee 100
+e
Aco011363.1 146 SE 147
87 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50891 | 26.946 | 47 | 148 | IPR004883 | Lateral organ boundaries, LOB |
| Pfam | PF03195 | 1.3E-43 | 48 | 145 | IPR004883 | Lateral organ boundaries, LOB |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009799 | Biological Process | specification of symmetry | ||||
| GO:0009944 | Biological Process | polarity specification of adaxial/abaxial axis | ||||
| GO:0009954 | Biological Process | proximal/distal pattern formation | ||||
| GO:0048441 | Biological Process | petal development | ||||
| GO:0005654 | Cellular Component | nucleoplasm | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 270 aa Download sequence Send to blast |
MASSSSAPIH VSSSSVAAAS VPSSSAAAAS AAAAAAAAAA AGGGTSSPCA ACKFLRRKCQ 60 PDCVFAPYFP PDNPQKFVHV HRVFGASNVT KLLNELHPYQ REDAVNSLAY EADMRLRDPV 120 YGCVGVISIL QHQLRQLQMD LTTAKSELSK YQAAAAAAAA AASAAPSTAN FIGGMSSGPP 180 GGSGAHTFMN MNHPIGIGIS SGGSGGFGRD QFFSVGMREQ QAMMRGYDDM TARVAAMSAV 240 GMGPLGGGGG VQFLRPSAAG GDERPSSGP* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5ly0_A | 8e-55 | 38 | 151 | 1 | 114 | LOB family transfactor Ramosa2.1 |
| 5ly0_B | 8e-55 | 38 | 151 | 1 | 114 | LOB family transfactor Ramosa2.1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Promotes the switch from proliferation to differentiation in the embryo sac. Negative regulator of cell proliferation in the adaxial side of leaves. Regulates the formation of a symmetric lamina and the establishment of venation. Interacts directly with RS2 (rough sheath 2) to repress some knox homeobox genes. {ECO:0000269|PubMed:17209126}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00211 | DAP | Transfer from AT1G65620 | Download |
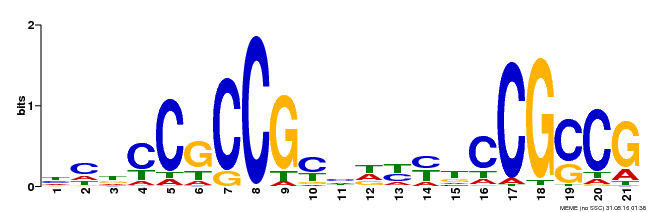
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC134933 | 2e-93 | AC134933.2 Oryza sativa Japonica Group cultivar Nipponbare chromosome 5 clone P0579A05, complete sequence. | |||
| GenBank | AC137617 | 2e-93 | AC137617.3 Oryza sativa Japonica Group cultivar Nipponbare chromosome 5 clone OSJNBa0084P24, complete sequence. | |||
| GenBank | AP014961 | 2e-93 | AP014961.1 Oryza sativa Japonica Group DNA, chromosome 5, cultivar: Nipponbare, complete sequence. | |||
| GenBank | AY940681 | 2e-93 | AY940681.1 Zea mays ASYMMETRIC LEAVES2 mRNA, complete cds. | |||
| GenBank | EF081454 | 2e-93 | EF081454.1 Zea mays indeterminate gametophyte 1 (IG1) gene, complete cds. | |||
| GenBank | EU964817 | 2e-93 | EU964817.1 Zea mays clone 281759 mRNA sequence. | |||
| GenBank | EU966092 | 2e-93 | EU966092.1 Zea mays clone 291632 mRNA sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020086921.1 | 0.0 | LOB domain-containing protein 6 | ||||
| Refseq | XP_020086930.1 | 0.0 | LOB domain-containing protein 6 | ||||
| Refseq | XP_020086938.1 | 0.0 | LOB domain-containing protein 6 | ||||
| Swissprot | Q32SG3 | 1e-85 | LBD6_MAIZE; LOB domain-containing protein 6 | ||||
| TrEMBL | A0A199VA05 | 1e-127 | A0A199VA05_ANACO; LOB domain-containing protein 6 | ||||
| STRING | XP_008807271.1 | 4e-92 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2267 | 38 | 95 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G65620.4 | 4e-56 | LBD family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aco011363.1 |



