 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aco012845.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Bromeliaceae; Ananas
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1033aa MW: 117208 Da PI: 7.3477 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 162 | 1.1e-50 | 123 | 235 | 5 | 117 |
CG-1 5 kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqrrcywlL 102
+rw +++e++aiL n+++++++ ++ ++p+sg+++L++rk++r+frkDG++wkkkkdgktv+E+hekLK+g+ve +++yYa+ e++p+f rr+ywlL
Aco012845.1 123 PNRWFRPNEVHAILSNYARFKIQPQPVDKPQSGTVLLFDRKMLRNFRKDGHNWKKKKDGKTVQEAHEKLKIGNVERIHVYYARCEDDPNFYRRSYWLL 220
68************************************************************************************************ PP
CG-1 103 eeelekivlvhylev 117
++ le+ivlvhy+++
Aco012845.1 221 DKMLERIVLVHYRQT 235
************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 72.915 | 115 | 241 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 1.7E-62 | 118 | 236 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 2.6E-44 | 123 | 234 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 5.6E-13 | 477 | 564 | IPR014756 | Immunoglobulin E-set |
| SuperFamily | SSF48403 | 3.11E-16 | 657 | 784 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 1.1E-15 | 667 | 784 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 4.02E-14 | 667 | 777 | No hit | No description |
| PROSITE profile | PS50297 | 16.255 | 685 | 789 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 4.4E-8 | 691 | 781 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 11.14 | 718 | 750 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 1.9E-5 | 718 | 747 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 260 | 757 | 786 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50096 | 6.998 | 870 | 896 | IPR000048 | IQ motif, EF-hand binding site |
| SuperFamily | SSF52540 | 2.42E-6 | 871 | 936 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 0.39 | 885 | 907 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.236 | 886 | 913 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0011 | 888 | 906 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 1.3 | 908 | 930 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.322 | 910 | 933 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0033 | 911 | 929 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 2.2 | 990 | 1012 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.919 | 992 | 1020 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.24 | 993 | 1012 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1033 aa Download sequence Send to blast |
MLHTLSLDTP KLEPSNLQLT VKIPRSVHRY PVYLTPRRRI TVHALLAPSP FLPALPLSFS 60 LSSDTKLRGR SYLPFLFSIT SSARVSPSSP FAAMEGGLLL DGEIHGFHTA SDLDVEKLME 120 DAPNRWFRPN EVHAILSNYA RFKIQPQPVD KPQSGTVLLF DRKMLRNFRK DGHNWKKKKD 180 GKTVQEAHEK LKIGNVERIH VYYARCEDDP NFYRRSYWLL DKMLERIVLV HYRQTSEDHA 240 SQYLPAPVDC TENLSLISKI HHDSPSTPIE SNSASAHSVV SGSAGTSEEI NSGEDNTVQA 300 ELQSHEFSLH EINTLDWDDL LESPTNNDST QHRDGDFFIS DQQIQNELAK SMNNGSFFMP 360 HNTSCITDND GAGNDQTIGS YFQAMKDQES QSQLLNSTKS ESQTEDRTSN AITGTSTPDN 420 ITNDVFLSQN SLGRWNYLND DINLLDDLQL EPLRSEANKL ATCTLMDQSS NQQPIFNITD 480 VSPGWAYSTE ETKVLVVGYF YEQQKHLMET QVYCGFGRKC VPAELVQAGV YRCTAAPSEP 540 GFVNLYLTLD GQTPISQVWS FEYRSKLSVW DEVASSEDDN RKLKLKEFQM QMRLAHLLFS 600 TSGNVSGLSG RTQQKFLKEA KNFASLSSSL MEKEWLNMLK LVNSNRDSQM PATEELLELV 660 LRNKLQEWLL EKVAEGRRTT PCDIQGQGVI HLCAILDYPW AVYLFSLSGF SLDFRDACGW 720 TALHWAAYYG RERMVATLLS AGANPSLVTD PTPESPGGST AADLAAKQGY EGLAAYLAEK 780 GLTAHFQAMS LSGNVATAAP FSTKKVNFES TDIENLSEQE LCLKESLAAY RNAADAADRI 840 QAAFRERSLK LQTKAVQMSK PEMEASEIVA ALKIQHAFRN HNRRKLMRAA AQIQSHFRTW 900 KTRKNFLHVR RQVIRIQAAF RGHQVRKQYC KIIWSVGVLE KAILRWRLKR RGLRGIHTEA 960 PSEAMRVDTE PANTEEEEFF RISREQAEER VKRSVIRVQT MFRSYRAQQE YRRMRMAHEK 1020 AKLEFNQMGH FY* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
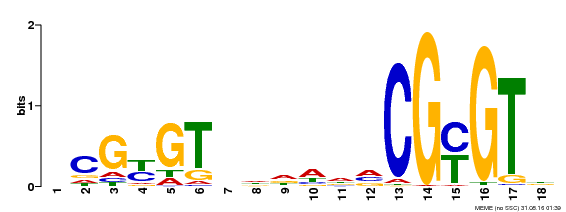
| UniProt | Transcription activator that binds calmodulin in a calcium-dependent manner in vitro. Binds to the DNA consensus sequence 5'-T[AC]CG[CT]GT[GT][GT][GT][GT]T[GT]CG-3'. {ECO:0000269|PubMed:16192280}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020084464.1 | 0.0 | calmodulin-binding transcription activator 5 isoform X2 | ||||
| Swissprot | Q7XHR2 | 0.0 | CBT_ORYSJ; Calmodulin-binding transcription activator CBT | ||||
| TrEMBL | A0A199W1X9 | 0.0 | A0A199W1X9_ANACO; Calmodulin-binding transcription activator 5 | ||||
| STRING | XP_008786234.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP9831 | 29 | 33 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aco012845.1 |



