 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aco015338.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Bromeliaceae; Ananas
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 384aa MW: 39394.4 Da PI: 8.9614 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 135 | 2.4e-42 | 99 | 174 | 1 | 76 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkk 76
+CqvegC+adls ak y+ rhkvC +hsk+p v+v+g+eqrfCqqCsrfh+l efD++krsCrrrLa+hnerrrk+
Aco015338.1 99 RCQVEGCKADLSGAKAYYCRHKVCGMHSKSPRVIVAGIEQRFCQQCSRFHQLPEFDQGKRSCRRRLAGHNERRRKP 174
6*************************************************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 1.5E-34 | 94 | 161 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.308 | 97 | 174 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 5.89E-40 | 98 | 177 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 1.3E-32 | 100 | 173 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0048653 | Biological Process | anther development | ||||
| GO:2000025 | Biological Process | regulation of leaf formation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 384 aa Download sequence Send to blast |
MEEMGLSSSD SLNGLKFGKK IYFEDGGGGG GGSGNGGGGG GGGGSGNGTA AAASASSTSK 60 APAPAAAAAP PGKKGKGASS AAAAAAAAAA AAAAAQVPRC QVEGCKADLS GAKAYYCRHK 120 VCGMHSKSPR VIVAGIEQRF CQQCSRFHQL PEFDQGKRSC RRRLAGHNER RRKPPSGPLS 180 SRYGRLALSF HEQPSRFRSF LMDFTYPNAL PSTGRDAWPT IRAGDRASNS HWQEGLNPNH 240 NSGTVTVNGA HAYIQGNLFN ASELPQGECL GGVGSESGCA LSLLSTTQPW GNHTTGNATP 300 TITLSSAGFD GSSTAPPMNV GNYETGPWGF RVHGTRSSSH EMPHEVGQFS GELELALQGN 360 GPHLDHGSGR GFEPSGHGMN WSL* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 1e-30 | 100 | 173 | 11 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 32 | 44 | SGNGGGGGGGGGS |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' (By similarity). May be involved in panicle development. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00307 | DAP | Transfer from AT2G42200 | Download |
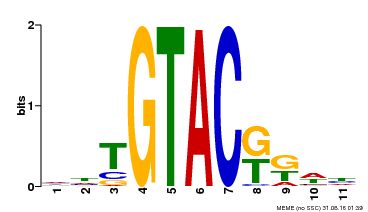
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Negatively regulated by microRNAs miR156b and miR156h. {ECO:0000305|PubMed:16861571}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AJ011619 | 5e-42 | AJ011619.1 Zea mays mRNA for SBP-domain protein 6, partial. | |||
| GenBank | BT041777 | 5e-42 | BT041777.2 Zea mays full-length cDNA clone ZM_BFb0053F21 mRNA, complete cds. | |||
| GenBank | EU972995 | 5e-42 | EU972995.1 Zea mays clone 391427 squamosa promoter-binding-like protein 9 mRNA, complete cds. | |||
| GenBank | HQ858679 | 5e-42 | HQ858679.1 Zea mays clone UT1135 SBP transcription factor mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020110587.1 | 0.0 | squamosa promoter-binding-like protein 14 | ||||
| Swissprot | Q7EXZ2 | 7e-93 | SPL14_ORYSJ; Squamosa promoter-binding-like protein 14 | ||||
| TrEMBL | A0A199UEX2 | 1e-174 | A0A199UEX2_ANACO; Squamosa promoter-binding-like protein 14 | ||||
| STRING | XP_008795751.1 | 1e-139 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2527 | 37 | 87 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G42200.1 | 8e-50 | squamosa promoter binding protein-like 9 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aco015338.1 |



