 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aco015663.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Bromeliaceae; Ananas
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 427aa MW: 47717.9 Da PI: 8.0655 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 34.4 | 4.7e-11 | 129 | 158 | 5 | 34 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeek 34
k rr+++NReAAr+sR+RKka+i++Le
Aco015663.1 129 KTLRRLAQNREAARKSRLRKKAYIQQLESS 158
7789***********************975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.20.5.170 | 1.3E-8 | 124 | 171 | No hit | No description |
| SMART | SM00338 | 1.1E-6 | 125 | 213 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 9.209 | 127 | 171 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 3.75E-7 | 129 | 171 | No hit | No description |
| Pfam | PF00170 | 4.2E-8 | 129 | 158 | IPR004827 | Basic-leucine zipper domain |
| PROSITE pattern | PS00036 | 0 | 132 | 147 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF14144 | 1.6E-30 | 222 | 296 | IPR025422 | Transcription factor TGA like domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 427 aa Download sequence Send to blast |
MIHPSSSNNI HGNIMRKDSG GYDFAELDQA LFLYLDGQEQ SGSQEHRQTL NIFPSQPMHV 60 EPSNSKGPIS LIPNTSSTSK GPSNQSMEPG KPRTETGKGT KPTVKREGNN GKATVSNSDQ 120 EGPKTPDPKT LRRLAQNREA ARKSRLRKKA YIQQLESSRI RLSQIEQELQ RSRAQGVLFG 180 GGASGGLLGE QGLPTSMGGL SSDAAMFDLE YGRWLEEHHK IMCQLRAALE EHLPENQLRM 240 FVESTLSHYE VLMNLKSIVA KSDVFHLFSG MWKTPTERCF LWIGDFRPSE LIKIVLRHIE 300 PLSEQQFLGI CGLQQSMQES EEALNQGLEA LHHSLSETIN SDVLSSPSNM ANYMGHMSVA 360 INKLTALEGF VRQADNLRQQ TLHRLQQILT TQQSARCFLE IAEYFHRLRA LSSLWIARPR 420 QDGPFG* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to as1-like elements (5'-TGACGTAAgggaTGACGCA-3') in promoters of target genes. Regulates transcription in response to plant signaling molecules salicylic acid (SA), methyl jasmonate (MJ) and auxin (2,4D) only in leaves. Prevents lateral branching and may repress defense signaling. {ECO:0000269|PubMed:12777042}. | |||||
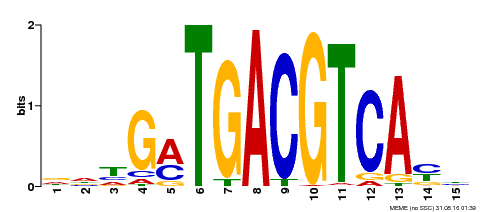
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00491 | DAP | Transfer from AT5G06839 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020109111.1 | 0.0 | transcription factor HBP-1b(c38)-like isoform X1 | ||||
| Swissprot | Q52MZ2 | 0.0 | TGA10_TOBAC; bZIP transcription factor TGA10 | ||||
| TrEMBL | A0A2H3YM29 | 0.0 | A0A2H3YM29_PHODC; bZIP transcription factor TGA10-like | ||||
| STRING | XP_008800656.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2732 | 36 | 69 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G06839.3 | 1e-165 | bZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aco015663.1 |



