 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aco018444.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Bromeliaceae; Ananas
|
||||||||
| Family | ARR-B | ||||||||
| Protein Properties | Length: 677aa MW: 74157.2 Da PI: 5.5098 | ||||||||
| Description | ARR-B family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 89.4 | 3.4e-28 | 233 | 286 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
kpr++W+ eLH++Fv+av++L G +kA+Pk+ilelm+v+gLt+e+v+SHLQk+Rl
Aco018444.1 233 KPRVVWSVELHQQFVHAVNHL-GFDKAVPKKILELMNVPGLTRENVASHLQKFRL 286
79*******************.********************************8 PP
| |||||||
| 2 | Response_reg | 73.1 | 1.1e-24 | 49 | 157 | 1 | 109 |
EEEESSSHHHHHHHHHHHHHTTCEEEEEESSHHHHHHHHHHHH..ESEEEEESSCTTSEHHHHHHHHHHHTTTSEEEEEESTTTHHHHHHHHHTTES CS
Response_reg 1 vlivdDeplvrellrqalekegyeevaeaddgeealellkekd..pDlillDiempgmdGlellkeireeepklpiivvtahgeeedalealkaGak 95
vl+vdD+++ + +l+q l + y +v++++++++al ll+e++ +D+++ D+ mp+mdG++ll+ e++lp+i+++a +++ + +k Ga
Aco018444.1 49 VLVVDDDDTCLTILKQKLLRCCY-DVTTCSEATKALSLLRESKggFDVVISDVHMPDMDGFRLLELVGL-EMDLPVIMMSADTRTSLVMKGIKHGAC 143
89*********************.******************99********************87754.558************************ PP
EEEESS--HHHHHH CS
Response_reg 96 dflsKpfdpeelvk 109
d+l Kp+ +eel +
Aco018444.1 144 DYLIKPVRIEELKN 157
***********987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF52172 | 3.84E-34 | 46 | 170 | IPR011006 | CheY-like superfamily |
| Gene3D | G3DSA:3.40.50.2300 | 3.2E-40 | 46 | 186 | No hit | No description |
| SMART | SM00448 | 3.6E-26 | 47 | 159 | IPR001789 | Signal transduction response regulator, receiver domain |
| PROSITE profile | PS50110 | 40.661 | 48 | 163 | IPR001789 | Signal transduction response regulator, receiver domain |
| Pfam | PF00072 | 1.3E-21 | 49 | 157 | IPR001789 | Signal transduction response regulator, receiver domain |
| CDD | cd00156 | 4.81E-24 | 50 | 163 | No hit | No description |
| PROSITE profile | PS51294 | 11.547 | 230 | 289 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.0E-29 | 230 | 291 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 6.45E-20 | 231 | 290 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 2.7E-24 | 233 | 286 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 7.3E-8 | 235 | 284 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009873 | Biological Process | ethylene-activated signaling pathway | ||||
| GO:0010082 | Biological Process | regulation of root meristem growth | ||||
| GO:0010119 | Biological Process | regulation of stomatal movement | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0071368 | Biological Process | cellular response to cytokinin stimulus | ||||
| GO:0080113 | Biological Process | regulation of seed growth | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 677 aa Download sequence Send to blast |
MIGFDFGVFE GCFGGWGIFW AALSGVFGKG VAMAEEWAEW DRFPVGLKVL VVDDDDTCLT 60 ILKQKLLRCC YDVTTCSEAT KALSLLRESK GGFDVVISDV HMPDMDGFRL LELVGLEMDL 120 PVIMMSADTR TSLVMKGIKH GACDYLIKPV RIEELKNIWQ HVFRKKQKEY EPSGGSLEDS 180 NRNNNKKACD DADYASLGKE GADGSSKSQK KRRDVKEEDD GELENSDPSS SKKPRVVWSV 240 ELHQQFVHAV NHLGFDKAVP KKILELMNVP GLTRENVASH LQKFRLYLKK MSEVSQQQTP 300 HHNSFSGLPS NVRANTLNQL DLQSWTVSSQ ITPQTLAAFQ NELLGQPSNN LILSGTDQPS 360 LRQVSVQAPN LESERVFSWP ILKSQPNLSR QLSQASAIPV EGISSAFPSW SCDDLSGQIT 420 NSRNSTTMSQ ILQQQQQGFD SFDLGKNSML INQNPVLPTS STNQDPNFSL SQSSASINAA 480 SSCGLLDQKF IIANTNVETR CDPFQMSQHS MKVTSQASSD LLSQNSSYNN SSIGIMDYSL 540 LMPQSKSVRF GVGQVVANSY SVPELDSSHV SSCIDDAIWG QLRNLDWNIN NLTSGSPSLV 600 PNSCNIQNSS GLLSNLPNPR QGRSSLGFVG KGTCIPSRFA INDVESPTNN LSHSNTCTDD 660 GGDMPNAVRF GLNGQL* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 5e-24 | 229 | 292 | 1 | 64 | ARR10-B |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that binds specific DNA sequence. Functions as a response regulator involved in His-to-Asp phosphorelay signal transduction system. Phosphorylation of the Asp residue in the receiver domain activates the ability of the protein to promote the transcription of target genes. May directly activate some type-A response regulators in response to cytokinins. {ECO:0000250|UniProtKB:Q940D0}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00054 | PBM | Transfer from AT4G16110 | Download |
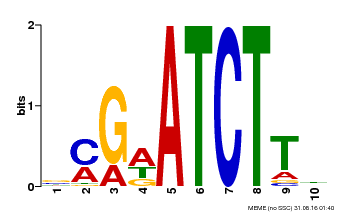
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KM661031 | 0.0 | KM661031.1 Ananas bracteatus clone 41159b microsatellite sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020105708.1 | 0.0 | two-component response regulator ORR21-like isoform X1 | ||||
| Swissprot | A2XE31 | 1e-168 | ORR21_ORYSI; Two-component response regulator ORR21 | ||||
| TrEMBL | A0A199VWM6 | 0.0 | A0A199VWM6_ANACO; Two-component response regulator | ||||
| STRING | XP_010271366.1 | 0.0 | (Nelumbo nucifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP5796 | 37 | 54 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16110.1 | 1e-104 | response regulator 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aco018444.1 |



