 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aco021664.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Bromeliaceae; Ananas
|
||||||||
| Family | TCP | ||||||||
| Protein Properties | Length: 356aa MW: 37415.6 Da PI: 5.618 | ||||||||
| Description | TCP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | TCP | 116.4 | 3.6e-36 | 59 | 166 | 7 | 100 |
TCP 7 rhskihTkvggRdRRvRlsaecaarfFdLqdeLGfdkdsktieWLlqqakpaikeltgtssssaseceaesssssasnsssg...............k 89
r+ ++hTkv+gR+RR+R++a caar+F+L++eLG+++d++ti WLlqqa+pai+++tgt++ +a ++ +++ + ++s++ +
Aco021664.1 59 RNRDRHTKVEGRGRRIRMPAVCAARIFQLTRELGHKSDGETIRWLLQQAEPAIIAATGTGTVPAIAT-TVDGTLQIPTQSAAaglegggggggggedS 155
899********************************************************99999555.444444444444444566677777776542 PP
TCP 90 aaksaakskks 100
aak +k +++
Aco021664.1 156 AAKRRKKLQPT 166
22222222222 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03634 | 9.9E-30 | 60 | 153 | IPR005333 | Transcription factor, TCP |
| PROSITE profile | PS51369 | 27.009 | 60 | 114 | IPR017887 | Transcription factor TCP subgroup |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0008361 | Biological Process | regulation of cell size | ||||
| GO:0048364 | Biological Process | root development | ||||
| GO:1900056 | Biological Process | negative regulation of leaf senescence | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 356 aa Download sequence Send to blast |
MELERQDKEE EEEEEEELLR TAAAVPMHRF IVPKPEPMEL FGSLQLIRKQ QQQQAGGGRN 60 RDRHTKVEGR GRRIRMPAVC AARIFQLTRE LGHKSDGETI RWLLQQAEPA IIAATGTGTV 120 PAIATTVDGT LQIPTQSAAA GLEGGGGGGG GGEDSAAKRR KKLQPTRAAA TTTTTPTPAM 180 PTPMPMPMPP AAAAAAAYYP IVADSIFQVG GGGGGGAISM SSGLAPIAAG PQGLLPVWVV 240 PSPGVAAGPS SQAQLWAIPT PPQIIGLAAP PPPEAQPPPS APSGGAAEKG AELQLMGCES 300 EERRRRGGGE EGGDHHHLHH HHHHHHHDHH HHHDDGPEEE EDDEEEPVSE SSNEE* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator. Binds the promoter core sequence 5'-GGNCC-3', especially at sites IIa (5'-GGGCCCAC-3') and IIb (5'-GGTCCCAC-3') (essential for meristematic tissue-specificity expression) of the PCNA gene promoter. {ECO:0000269|PubMed:12000681}. | |||||
| UniProt | Transcription activator. Binds the promoter core sequence 5'-GGNCC-3', especially at sites IIa (5'-GGGCCCAC-3') and IIb (5'-GGTCCCAC-3') (essential for meristematic tissue-specificity expression) of the PCNA gene promoter. {ECO:0000269|PubMed:12000681, ECO:0000269|PubMed:9338963}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00667 | PBM | Transfer from GSVIVG01027588001 | Download |
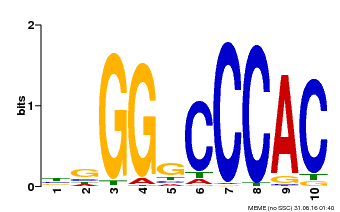
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EU945866 | 1e-58 | EU945866.1 Zea mays clone 288123 mRNA sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020091451.1 | 0.0 | transcription factor PCF2-like | ||||
| Swissprot | A2YXQ1 | 3e-76 | PCF2_ORYSI; Transcription factor PCF2 | ||||
| Swissprot | Q6ZBH6 | 3e-76 | PCF2_ORYSJ; Transcription factor PCF2 | ||||
| TrEMBL | A0A199W885 | 0.0 | A0A199W885_ANACO; Transcription factor PCF2 | ||||
| STRING | XP_008809784.1 | 3e-81 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3912 | 31 | 67 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G45680.1 | 1e-44 | TCP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aco021664.1 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



