 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aco022096.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Bromeliaceae; Ananas
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 665aa MW: 71906.4 Da PI: 6.2309 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 50.8 | 2.9e-16 | 389 | 435 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K++Ka++L +A+eY+k Lq
Aco022096.1 389 VHNLSERRRRDRINEKMRALQELIPNC-----NKVDKASMLDEAIEYLKTLQ 435
6*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 9.4E-21 | 383 | 443 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.31E-20 | 383 | 446 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.349 | 385 | 434 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 2.23E-17 | 388 | 439 | No hit | No description |
| Pfam | PF00010 | 9.7E-14 | 389 | 435 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 7.6E-18 | 391 | 440 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 665 aa Download sequence Send to blast |
MTNNSSSDRS SFMPDNDFVE LLWEDGQIVM QGQSSRPKRS FVPTPFNPSY NNRFQDKDIK 60 NEILPKQLNH FESGDPTVRH DFFTSDPLND KDDDDDDSVP WINYPMVDDS LSNDFCSEFL 120 AEFSGLNPNN VSISHNTDQI VRGSSSMENN HAPSKAVIGG SGGFTSRSGQ LLESSQQHLN 180 SGHAVKSKAT DFGTASSIVT HSRMPQPSGT SPLLNFSHFS RPVSLAKANL ESMERLRSNE 240 KAFTAPTSSN PPESTLIQST GGFKELRPSS ANTPKEMVSL EHTQALCQQE MPRKNNNHTA 300 VANSNCINQQ GSGFAPQKGP ETIVASSSVC SGNSAGAASD DPKHGLKRKD CEGEDSGNHS 360 EDVEDESVGL KRPANVRATK AKRTRAAEVH NLSERRRRDR INEKMRALQE LIPNCNKVDK 420 ASMLDEAIEY LKTLQLQVQM MSMGGSLCMP PMMLPPGMQQ MHLPSMAPHY AQMGVGMGLN 480 VRLGYGIGPL DMSSSPNCPL IPVPPIHCGT QFPCPTVPGA QVRPAMAGPG NLPMFAIPGL 540 GAIPSAVPRI VPQLGSFSGL AVMANPVPVP ASDTTTPLNC KEHREHSMNL ERKKSSSDSQ 600 KTPFAPSALA QSNQTLHSSG SGGANSDCSL NFEWPYDLHE HDGLGFDFCA ICAKAVQAED 660 FGMLG |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 393 | 398 | ERRRRD |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
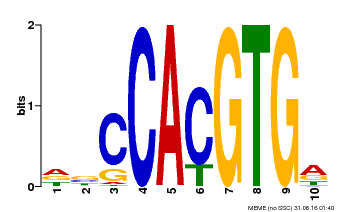
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020086865.1 | 0.0 | transcription factor PIF3-like isoform X1 | ||||
| Refseq | XP_020086866.1 | 0.0 | transcription factor PIF3-like isoform X1 | ||||
| Refseq | XP_020086867.1 | 0.0 | transcription factor PIF3-like isoform X1 | ||||
| Refseq | XP_020086868.1 | 0.0 | transcription factor PIF3-like isoform X1 | ||||
| Refseq | XP_020086869.1 | 0.0 | transcription factor PIF3-like isoform X1 | ||||
| TrEMBL | A0A199V921 | 0.0 | A0A199V921_ANACO; Transcription factor PIF3 | ||||
| STRING | XP_008800313.1 | 1e-158 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3527 | 37 | 75 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 5e-41 | phytochrome interacting factor 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aco022096.1 |



