 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aco022888.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Bromeliaceae; Ananas
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 566aa MW: 59148.8 Da PI: 6.587 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 97.2 | 1.1e-30 | 293 | 351 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-ST.T---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsa.gCpvkkkversaedpkvveitYegeHnhe 59
Dg++WrKYGqK+ kg+++pr+YYrCt+a gCpv+k+v+r+aed +++++tYeg+Hnh+
Aco022888.1 293 TDGCQWRKYGQKMAKGNPCPRAYYRCTMAtGCPVRKQVQRCAEDRSILITTYEGNHNHP 351
7***************************99****************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 1.1E-33 | 277 | 353 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 6.41E-28 | 285 | 353 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 29.631 | 287 | 353 | IPR003657 | WRKY domain |
| SMART | SM00774 | 3.1E-37 | 292 | 352 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 6.4E-27 | 294 | 351 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0016036 | Biological Process | cellular response to phosphate starvation | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0080169 | Biological Process | cellular response to boron-containing substance deprivation | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 566 aa Download sequence Send to blast |
MDKPGGGGGG GGGGLTSNAN SASLGFFTAA DPGRIFFHSR PHSMDTGSFD PPKSVEMDFF 60 SNEKKKSSRG AAAEPDLGLK VPGLAIKKED LTINTGLQLL TANTKSDQST VDDGIYPNEE 120 DKEEKSELAA MQAELGRMNE ENQKLRGMLN QVTSNYNALQ MHLFTLLQQR NQKNVGNQSS 180 HEAVDEKMES GAMVPRQFMD LGPAVADHAT DEASNSSTEG GSLDRSTSPT TNKEVAAIDN 240 ERAAREDSPD DGWIPNKNPK MAPPKGPEQQ AQEATMRKAR VSVRARSEAP MITDGCQWRK 300 YGQKMAKGNP CPRAYYRCTM ATGCPVRKQV QRCAEDRSIL ITTYEGNHNH PLPPAAMAMA 360 STTSAAATML LSGSMSSSAG LMNPNFLART ILPCSTSMAT ISASAPFPTV TLDLTQTPNN 420 QLHQLQRPLA QFGVPLIPSG AAAGPAAPFG AAIPQVIGQA LYNQSKLAGL HMSGDAVPTA 480 VAPPASLADT VTAATAAITA DPNFTAALAA AIASIIGGAH QTGNNNNANN NNAINHNSNN 540 NNNNNSTSSG SGGGGVPTNS NFPAT* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2ayd_A | 4e-23 | 279 | 355 | 1 | 76 | WRKY transcription factor 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in the control of processes related to senescence and pathogen defense (PubMed:12000796). Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (PubMed:12000796). Activates the transcription of the SIRK gene and represses its own expression and that of the WRKY42 genes (PubMed:12000796). Modulates phosphate homeostasis and Pi translocation by regulating PHO1 expression (PubMed:25733771). {ECO:0000269|PubMed:12000796, ECO:0000269|PubMed:25733771}. | |||||
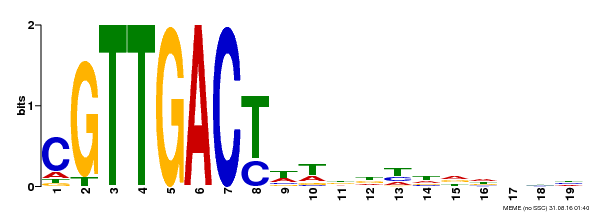
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00209 | DAP | Transfer from AT1G62300 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salicylic acid, ethylene, jasmonic acid, pathogens, wounding and strongly during leaf senescence. {ECO:0000269|PubMed:11449049, ECO:0000269|PubMed:11722756}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020100979.1 | 0.0 | probable WRKY transcription factor 31 | ||||
| Swissprot | Q9C519 | 1e-146 | WRKY6_ARATH; WRKY transcription factor 6 | ||||
| TrEMBL | A0A199W4G7 | 0.0 | A0A199W4G7_ANACO; WRKY transcription factor 6 | ||||
| STRING | XP_008784385.1 | 0.0 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP5265 | 36 | 56 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G62300.1 | 1e-121 | WRKY family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aco022888.1 |



