 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aco031477.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Bromeliaceae; Ananas
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 330aa MW: 35544.6 Da PI: 10.6637 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 99.8 | 1.7e-31 | 233 | 291 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-ST.T---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsa.gCpvkkkversaedpkvveitYegeHnhe 59
D+y+WrKYGqK++kgs++pr+YY+C+s gCp++k+ver+ +dp+v+++tYegeH+h+
Aco031477.1 233 ADEYSWRKYGQKPIKGSPYPRGYYKCSSLrGCPARKHVERAPDDPSVLIVTYEGEHRHS 291
69*************************998****************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF10533 | 1.5E-17 | 176 | 230 | IPR018872 | Zn-cluster domain |
| Gene3D | G3DSA:2.20.25.80 | 3.0E-33 | 220 | 291 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 30.366 | 227 | 293 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.44E-26 | 229 | 292 | IPR003657 | WRKY domain |
| SMART | SM00774 | 8.9E-38 | 232 | 292 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 8.4E-27 | 234 | 290 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009938 | Biological Process | negative regulation of gibberellic acid mediated signaling pathway | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:1990841 | Molecular Function | promoter-specific chromatin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 330 aa Download sequence Send to blast |
MAVDLMGHAK MNEQMVIQEA AAAGIRGMER LILQISHHEE PSPSPSPSSQ MQMDCREVTE 60 LTVSKFKNLI ALLNRTGHAR FRRGPPAPPP PPPPPPRPQA LTLAPPPPKA RTLDFARPAM 120 SSVSAPMSSS SSSFLSSVTA GDGSVSNGGQ QQPVGPLALR LVPAGWKPPL SVAPLKRKCD 180 AHAHAHAHAH SEDVAGKYGV NGGRCHCSKR RKLRVRKTIR VPAISAKAAD IPADEYSWRK 240 YGQKPIKGSP YPRGYYKCSS LRGCPARKHV ERAPDDPSVL IVTYEGEHRH SLNPQHNHNP 300 NLLLLSSRSN NSSPSAPTAP LTIDSFRPA* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2ayd_A | 5e-22 | 220 | 294 | 2 | 75 | WRKY transcription factor 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (By similarity). Regulates rhizobacterium B.cereus AR156-induced systemic resistance (ISR) to P.syringae pv. tomato DC3000, probably by activating the jasmonic acid (JA)- signaling pathway (PubMed:26433201). {ECO:0000250, ECO:0000269|PubMed:26433201}. | |||||
| UniProt | Transcription factor. Interacts, when in complex with WRKY71, specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element. Represses specifically gibberellic acid (GA)-induced promoters in aleurone cells, probably by interfering with GAM1. {ECO:0000269|PubMed:16623886}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00464 | DAP | Transfer from AT4G31550 | Download |
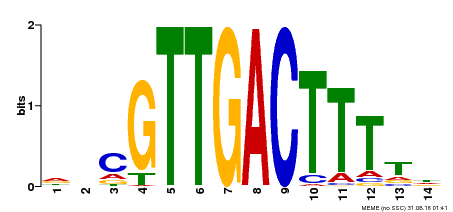
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by abscisic acid (ABA) in aleurone cells (PubMed:15618416, PubMed:16623886). Accumulates in response to uniconazole, a gibberellic acid (GA) biosynthesis inhibitor (PubMed:16623886). Repressed by GA (PubMed:15618416). {ECO:0000269|PubMed:15618416, ECO:0000269|PubMed:16623886}. | |||||
| UniProt | INDUCTION: Strongly during leaf senescence (PubMed:11722756). Repressed by rhizobacterium B.cereus AR156 in leaves, and to a lower extent, by P.fluorescens WCS417r (PubMed:26433201). {ECO:0000269|PubMed:11722756, ECO:0000269|PubMed:26433201}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020081192.1 | 0.0 | WRKY transcription factor WRKY51-like | ||||
| Swissprot | Q0JEE2 | 7e-96 | WRK51_ORYSJ; WRKY transcription factor WRKY51 | ||||
| Swissprot | Q9SV15 | 7e-96 | WRK11_ARATH; Probable WRKY transcription factor 11 | ||||
| TrEMBL | A0A199UUD8 | 1e-173 | A0A199UUD8_ANACO; Putative WRKY transcription factor 11 | ||||
| STRING | XP_008801524.1 | 1e-113 | (Phoenix dactylifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP617 | 38 | 171 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G31550.1 | 3e-57 | WRKY DNA-binding protein 11 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aco031477.1 |



