 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aqcoe1G083200.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Ranunculales; Ranunculaceae; Thalictroideae; Aquilegia
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 302aa MW: 34195.4 Da PI: 8.9258 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 28.6 | 3.3e-09 | 25 | 70 | 3 | 46 |
SS-HHHHHHHHHHHHHTTTT...-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGgg...tWktIartmgkgRtlkqcksrwqk 46
+W++eE +l+ +a + + ++ +W ++a +++ g+t ++ + +
Aqcoe1G083200.1.p 25 KWSQEENKLFENALAIYDKDtpdRWQKVATMIP-GKTVMDVMNQYKE 70
8********************************.****999999875 PP
| |||||||
| 2 | Myb_DNA-binding | 44.7 | 3.1e-14 | 129 | 173 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT+eE++l++ + k++G+g+W+ I+r + +Rt+ q+ s+ qky
Aqcoe1G083200.1.p 129 PWTEEEHKLFLLGLKKYGKGDWRNISRNFVITRTPTQVASHAQKY 173
8*****************************99************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 2.3E-5 | 19 | 73 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51293 | 9.782 | 21 | 75 | IPR017884 | SANT domain |
| SMART | SM00717 | 1.3E-10 | 22 | 74 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 4.93E-13 | 24 | 79 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 8.7E-8 | 24 | 70 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 7.36E-9 | 25 | 72 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 2.1E-12 | 120 | 178 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 20.486 | 122 | 178 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 8.22E-17 | 124 | 177 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.5E-17 | 125 | 177 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 5.3E-13 | 126 | 176 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.09E-11 | 129 | 174 | No hit | No description |
| Pfam | PF00249 | 1.1E-12 | 129 | 173 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 302 aa Download sequence Send to blast |
MMKWGMEILN PHTNWLLEEN KMTTKWSQEE NKLFENALAI YDKDTPDRWQ KVATMIPGKT 60 VMDVMNQYKE LVNDVSDIEA GLIPIPGYTT SSFTLDWASN RTFAQSFDSN GKRGSSGRPS 120 DQERKKGVPW TEEEHKLFLL GLKKYGKGDW RNISRNFVIT RTPTQVASHA QKYFIRQLSG 180 GKDKRRSSIH DITTVSLTDN RPTSPDNSTS PPMDQTMLQQ HSELSGTPRA LFDWNQPHDG 240 SNMVFNSTSA NMFVPPSFGV NSYGMKMEGG QDLHRGIIHA SQFGPHNTFF QMQSSQCHPR 300 G* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2cjj_A | 1e-16 | 26 | 91 | 11 | 76 | RADIALIS |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Involved in the dorsovental asymmetry of flowers. Promotes ventral identity. {ECO:0000269|PubMed:11937495, ECO:0000269|PubMed:9118809}. | |||||
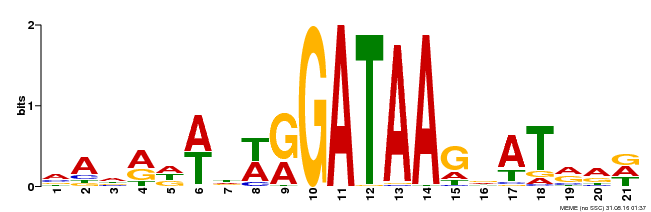
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00296 | DAP | Transfer from AT2G38090 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | FJ805376 | 1e-113 | FJ805376.1 Aquilegia alpina DIV protein (DIV) gene, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_026425763.1 | 1e-152 | transcription factor DIVARICATA-like | ||||
| Swissprot | Q8S9H7 | 1e-122 | DIV_ANTMA; Transcription factor DIVARICATA | ||||
| TrEMBL | A0A2G5EF51 | 0.0 | A0A2G5EF51_AQUCA; Uncharacterized protein | ||||
| STRING | Aquca_009_00736.1 | 0.0 | (Aquilegia coerulea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G38090.1 | 5e-91 | MYB family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aqcoe1G083200.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



