 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aqcoe1G159900.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Ranunculales; Ranunculaceae; Thalictroideae; Aquilegia
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 337aa MW: 38319 Da PI: 6.3657 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 54.7 | 2.3e-17 | 13 | 60 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg W++eEde+lv++++ +G +W+++++ g+ R++k+c++rw +yl
Aqcoe1G159900.1.p 13 RGLWSPEEDEKLVKCINTHGHSCWSAVPKLAGLQRCGKSCRLRWINYL 60
788*******************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 51.3 | 2.7e-16 | 66 | 109 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
rg+++++E+ +++d+++ lG++ W+ Ia++++ gRt++++k++w++
Aqcoe1G159900.1.p 66 RGSFSAQEERIIIDVHRILGNR-WAQIAKHLP-GRTDNEVKNFWNS 109
899*******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 2.6E-25 | 4 | 63 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 16.984 | 8 | 60 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.12E-28 | 10 | 107 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 9.4E-14 | 12 | 62 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 6.5E-16 | 13 | 60 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.30E-11 | 16 | 60 | No hit | No description |
| PROSITE profile | PS51294 | 25.757 | 61 | 115 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 6.1E-25 | 64 | 115 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 8.8E-16 | 65 | 113 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.0E-14 | 66 | 109 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.62E-11 | 68 | 108 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 337 aa Download sequence Send to blast |
MGHHCCSKQK VKRGLWSPEE DEKLVKCINT HGHSCWSAVP KLAGLQRCGK SCRLRWINYL 60 RPDLKRGSFS AQEERIIIDV HRILGNRWAQ IAKHLPGRTD NEVKNFWNSC IKKKLLAQGL 120 DPKTHNLMPP SQQNIKNACN VNQFYQQPPS VFSMNSQFRS DNNVTPMDIN QSFITLAPPH 180 PPNTNIYPPS LDQAMSTFNY QNQNHVWTTN AQNSEPSSDF TRQSLIPISS SSMIGSSGFD 240 FPDHENNNMW DDGGMEALVS PRQEEMKQTP VPQGLTQQEK VCEVQLEEER DGHHQDMDTS 300 FSDCSNFDLE FMESPLMPCG LMYSNMNSMD QLAWDA* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 5e-31 | 13 | 115 | 7 | 108 | B-MYB |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that regulates lignified secondary cell wall thickening of the anther endocethium, which is necessary for anther dehiscence (PubMed:12753590, PubMed:17147638, PubMed:17329564). May play a role in specifying early endothecial cell development by regulating a number of genes linked to secondary thickening such as NST1 and NST2. Acts upstream of the lignin biosynthesis pathway (PubMed:17329564). {ECO:0000269|PubMed:12753590, ECO:0000269|PubMed:17147638, ECO:0000269|PubMed:17329564}. | |||||
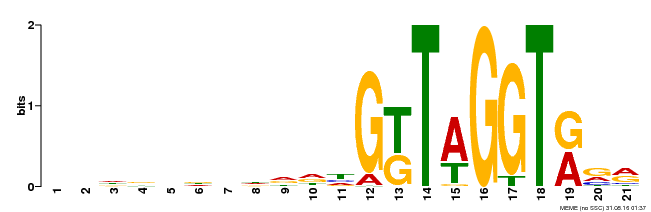
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00350 | DAP | Transfer from AT3G12720 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated by auxin. {ECO:0000269|PubMed:23410518}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016544702.1 | 1e-114 | PREDICTED: myb-related protein 330-like | ||||
| Refseq | XP_024462561.1 | 1e-114 | transcription factor MYB86 | ||||
| Swissprot | Q9SPG3 | 3e-66 | MYB26_ARATH; Transcription factor MYB26 | ||||
| TrEMBL | A0A2G5D2B3 | 0.0 | A0A2G5D2B3_AQUCA; Uncharacterized protein | ||||
| STRING | Aquca_030_00294.1 | 0.0 | (Aquilegia coerulea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G12720.1 | 5e-83 | myb domain protein 67 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aqcoe1G159900.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



