 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aqcoe1G199600.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Ranunculales; Ranunculaceae; Thalictroideae; Aquilegia
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 329aa MW: 36520.6 Da PI: 7.3796 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 105.3 | 3.2e-33 | 184 | 241 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
+Dgy+WrKYGqK vk+s++prsYYrCts++C vkk+vers ++p++v++tYeg+Hnh+
Aqcoe1G199600.1.p 184 EDGYRWRKYGQKAVKNSPYPRSYYRCTSQKCIVKKRVERSFQNPSIVITTYEGQHNHH 241
8********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 5.7E-35 | 168 | 243 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 4.05E-29 | 175 | 243 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 31.835 | 178 | 243 | IPR003657 | WRKY domain |
| SMART | SM00774 | 2.9E-38 | 183 | 242 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.7E-25 | 184 | 241 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 329 aa Download sequence Send to blast |
MADHRTRDLY NLNIYQGDHA GINGTSNFPI LTTTSPFNQI PSTEPSVEYQ QHGFDPMNMS 60 FTDYLQGSMD YGALERAFDL SRSSSEVFSS IDGSGGQNTG GGVGETVGSG DISAVTPNSS 120 ISSSSTEAAG EEDSGKSKKN RQEKGCEDGS EMSKKVNKPK KKPEKRPREP RVAFMTESEV 180 DHLEDGYRWR KYGQKAVKNS PYPRSYYRCT SQKCIVKKRV ERSFQNPSIV ITTYEGQHNH 240 HSPATLRGNA SRMLVTPMLT SPTMGPSFHQ ELLMQVPHTS NLLSDTSSTI TTMPPHNLTT 300 LQQLQIPDYG LLQDMVPPFM HRQQQQQQ* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 1e-27 | 173 | 244 | 7 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 1e-27 | 173 | 244 | 7 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
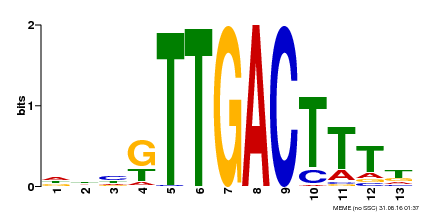
| UniProt | Transcription factor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00440 | DAP | Transfer from AT4G18170 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010275648.1 | 1e-106 | PREDICTED: probable WRKY transcription factor 71 | ||||
| Swissprot | Q8VWJ2 | 9e-65 | WRK28_ARATH; WRKY transcription factor 28 | ||||
| TrEMBL | A0A2G5DDD6 | 0.0 | A0A2G5DDD6_AQUCA; Uncharacterized protein | ||||
| STRING | Aquca_022_00127.1 | 0.0 | (Aquilegia coerulea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G18170.1 | 2e-55 | WRKY DNA-binding protein 28 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aqcoe1G199600.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



