 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aqcoe1G392900.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Ranunculales; Ranunculaceae; Thalictroideae; Aquilegia
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 593aa MW: 65536.3 Da PI: 7.7243 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 105.6 | 2.6e-33 | 250 | 306 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
dDgynWrKYGqK+vkgse+prsYY+Ct+++Cp+kkkvers d++++ei+Y+g+Hnh
Aqcoe1G392900.1.p 250 DDGYNWRKYGQKQVKGSENPRSYYKCTFPNCPTKKKVERSL-DGQITEIVYKGSHNHA 306
8****************************************.***************7 PP
| |||||||
| 2 | WRKY | 105.1 | 3.6e-33 | 419 | 477 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+vkg+++prsYY+Ct agCpv+k+ver+++d ++v++tYeg+Hnh+
Aqcoe1G392900.1.p 419 LDDGYRWRKYGQKVVKGNPNPRSYYKCTNAGCPVRKHVERASHDLRAVITTYEGKHNHD 477
59********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 3.2E-29 | 239 | 308 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.96E-25 | 243 | 308 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 23.376 | 244 | 308 | IPR003657 | WRKY domain |
| SMART | SM00774 | 2.4E-36 | 249 | 307 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 3.0E-25 | 250 | 305 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 2.9E-37 | 404 | 479 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 4.45E-29 | 411 | 479 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 38.572 | 414 | 479 | IPR003657 | WRKY domain |
| SMART | SM00774 | 7.7E-39 | 419 | 478 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 8.8E-26 | 420 | 477 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0009788 | Biological Process | negative regulation of abscisic acid-activated signaling pathway | ||||
| GO:0009938 | Biological Process | negative regulation of gibberellic acid mediated signaling pathway | ||||
| GO:0010120 | Biological Process | camalexin biosynthetic process | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0010508 | Biological Process | positive regulation of autophagy | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0070370 | Biological Process | cellular heat acclimation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 593 aa Download sequence Send to blast |
MASSTGSLDT SANSHPYNPH HQTFSFSNQY MNSSFSDLLS TGTGDDDENN RIMLRRSGGS 60 DGGLSDRIAD RTGSGIPKFK SIPPPSLPIS PPSVSPSSYF SIPPGLSPTE LLDSPVLLSS 120 SNLLPSPTTG TFPTQAFNWR ANSNSFQQGV KQEDRNYSDF SFQPQTKSSS GSIFQSSMSA 180 GDMYKGQQQQ QQQQQQPWSF QEPTKQSDFS TGKPIVKPET ASLQNNGAPQ SDYNQYAQPT 240 QTLREQRRSD DGYNWRKYGQ KQVKGSENPR SYYKCTFPNC PTKKKVERSL DGQITEIVYK 300 GSHNHAKPQS TRRSSASSQS IQAAVVPSSS EVSDHFGSHG NTQMDSVTTP ENSSISIGDE 360 DLDRSSQRSR SGGDEFDEDE PEAKKWKKES ENEVISASGS RTVREPRVVV QTTSDIDILD 420 DGYRWRKYGQ KVVKGNPNPR SYYKCTNAGC PVRKHVERAS HDLRAVITTY EGKHNHDVPA 480 ARGSGNHAVN RPLPDNSNHN NATMPIRPSA TTTQSNHNAN NTFRNGRLPT SEGQTPFTLE 540 MLQAPAGGYG FSSGYNNSMN SFMNQQQQTD NNVFPRTKEE PRDDMFMESL LY* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 2e-36 | 240 | 480 | 8 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 2e-36 | 240 | 480 | 8 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription repressor (By similarity). Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element. Negative regulator of both gibberellic acid (GA) and abscisic acid (ABA) signaling in aleurone cells, probably by interfering with GAM1, via the specific repression of GA- and ABA-induced promoters (By similarity). {ECO:0000250|UniProtKB:Q6IEQ7, ECO:0000250|UniProtKB:Q6QHD1}. | |||||
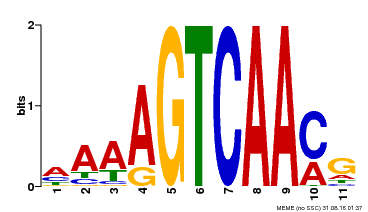
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00299 | DAP | Transfer from AT2G38470 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by abscisic acid (ABA) in aleurone cells, embryos, roots and leaves (PubMed:25110688). Slightly down-regulated by gibberellic acid (GA) (By similarity). Accumulates in response to jasmonic acid (MeJA) (PubMed:16919842). {ECO:0000250|UniProtKB:Q6IEQ7, ECO:0000269|PubMed:16919842, ECO:0000269|PubMed:25110688}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_023888322.1 | 0.0 | WRKY transcription factor WRKY24-like | ||||
| Swissprot | Q6B6R4 | 1e-144 | WRK24_ORYSI; WRKY transcription factor WRKY24 | ||||
| TrEMBL | A0A2G5EQU3 | 0.0 | A0A2G5EQU3_AQUCA; Uncharacterized protein | ||||
| STRING | Aquca_005_00209.1 | 0.0 | (Aquilegia coerulea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G38470.1 | 1e-109 | WRKY DNA-binding protein 33 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aqcoe1G392900.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



