 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aqcoe1G487700.3.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Ranunculales; Ranunculaceae; Thalictroideae; Aquilegia
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 556aa MW: 62628.7 Da PI: 6.9315 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 53.2 | 7.3e-17 | 233 | 282 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV++++++grW+++I+d + k+++lg f+ta Aa+a+++a+ k++g
Aqcoe1G487700.3.p 233 SQYRGVTFYRRTGRWESHIWD------CgKQVYLGGFDTAHAAARAYDRAAIKFRG 282
78*******************......55************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 6.7E-9 | 233 | 282 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 7.19E-16 | 233 | 291 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 1.3E-32 | 234 | 296 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 17.794 | 234 | 290 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 4.2E-17 | 234 | 291 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 2.50E-11 | 234 | 292 | No hit | No description |
| CDD | cd00018 | 3.35E-13 | 329 | 374 | No hit | No description |
| SuperFamily | SSF54171 | 2.55E-6 | 329 | 373 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 1.8E-13 | 330 | 378 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 556 aa Download sequence Send to blast |
MLDLNVISAE TIHRKEEEEE VDVISLNKKK KMMMLMMMNN NQNSNKDDLS AGTSNSSSVV 60 NAEVSSSNGA DEDSCSTHHH NNNNKFVVTT TTTATSSNST SVTWKEKDVF AIEFDILKKE 120 DFSREIETIV VDDEEEEDRN RNRNRNRNLI LQSDFVTKQF FPMTCVDELD LSLPVATMKA 180 ALSRTQSMDL CFTRSNVVGD LSEIRVLEQQ QQQQQQQKPP VKKSRRGPRS RSSQYRGVTF 240 YRRTGRWESH IWDCGKQVYL GGFDTAHAAA RAYDRAAIKF RGVDADINFN LSDYEEDMKQ 300 FDQQMKNLTK EEFVHILRRH STGFSRGSSK YRGVTLHKCG RWEARMGQFL GKKAYDKAAI 360 KCNGREAVTN FEPSTYDSEI MEIENGGNAC NLDLNLGITP PSSADDLNQS ESVGAFNLHV 420 DPLNLLDAKR ARTDNPSLTT LGGQPLYGLA MGSKHPPMWN GGYPSFFPNY EQERKIEKRV 480 EVGSRRLSNW AWQLHGHIDD STPMPLFSTA ASSGFSSSST SVSAAALHPP HPPHISMTHY 540 GYYPPSTTSQ HHYRS* |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00615 | PBM | Transfer from AT2G28550 | Download |
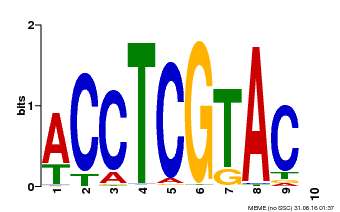
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010260158.1 | 1e-161 | PREDICTED: ethylene-responsive transcription factor RAP2-7-like isoform X2 | ||||
| TrEMBL | A0A2G5D5B4 | 0.0 | A0A2G5D5B4_AQUCA; Uncharacterized protein | ||||
| STRING | Aquca_027_00133.1 | 0.0 | (Aquilegia coerulea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G28550.1 | 9e-89 | related to AP2.7 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aqcoe1G487700.3.p |



