 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aqcoe2G014200.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Ranunculales; Ranunculaceae; Thalictroideae; Aquilegia
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 319aa MW: 35699.8 Da PI: 5.0453 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 45.2 | 2.2e-14 | 21 | 68 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g W++eEd++l ++ G g+W+ +ar g+ R++k+c++rw +yl
Aqcoe2G014200.1.p 21 KGLWSPEEDDKLMNYMLSNGQGCWSDVARNAGLQRCGKSCRLRWINYL 68
678*******************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 51.1 | 3.2e-16 | 74 | 117 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
rg+++++E+el+++++ lG++ W+ Ia++++ gRt++++k++w++
Aqcoe2G014200.1.p 74 RGAFSPQEEELIIHLHSVLGNR-WSQIAARLP-GRTDNEIKNFWNS 117
89********************.*********.***********97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 1.7E-22 | 13 | 71 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 16.223 | 16 | 68 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 6.64E-28 | 19 | 115 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 6.5E-10 | 20 | 70 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 4.1E-13 | 21 | 68 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.38E-8 | 24 | 68 | No hit | No description |
| PROSITE profile | PS51294 | 25.581 | 69 | 123 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.2E-26 | 72 | 124 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.5E-15 | 73 | 121 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 6.9E-15 | 74 | 117 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 7.64E-11 | 76 | 116 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:2000652 | Biological Process | regulation of secondary cell wall biogenesis | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 319 aa Download sequence Send to blast |
MRKPDPPGKN NMNTNNVKLR KGLWSPEEDD KLMNYMLSNG QGCWSDVARN AGLQRCGKSC 60 RLRWINYLRP DLKRGAFSPQ EEELIIHLHS VLGNRWSQIA ARLPGRTDNE IKNFWNSTIK 120 KRLKNSSTST SPNKSNASDP RDFTGGLFSM SEQDIMNLCM DSSSSSSSSM HAMVMRNRFD 180 PLPMLENSYD HMPNAGAGLF NVSACMTQVG IGDGFYGNHG IIGGGNIGLD QSELFIPSLE 240 SISIEENNIT DNTFEKTNNN NLLNNINIIN NNNNNNNIDN IVESGNYWGG ENLRMGEWNM 300 EELLEDVSSF PFLDFQVQ* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 2e-27 | 19 | 123 | 5 | 108 | B-MYB |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that acts as molecular switch in the NAC012/SND1-mediated transcriptional network regulating secondary wall biosynthesis. Is directly activated by NAC012/SND1 and its close homologs, including NAC043/NST1, NAC066/NST2, NAC101/VND6 and NAC030/VND7. Is required for functional expression of a number of secondary wall-associated transcription factors and secondary wall biosynthetic genes involved in cellulose, xylan and lignin synthesis. Functions redundantly with MYB46 in the transcriptional regulatory cascade leading to secondary wall formation in fibers and vessels (PubMed:19808805). Transcription activator that binds to the DNA consensus sequence 5'-ACC[AT]A[AC][TC]-3', designated as the secondary wall MYB-responsive element (SMRE). Regulates directly numerous transcription factors and a number of genes involved in secondary wall biosynthesis that contain SMRE elements in their promoters (PubMed:22197883). {ECO:0000269|PubMed:19808805, ECO:0000269|PubMed:22197883}. | |||||
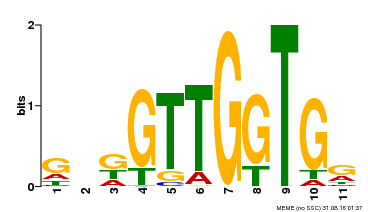
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00336 | DAP | Transfer from AT3G08500 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_022759455.1 | 1e-138 | transcription factor MYB83-like | ||||
| Swissprot | Q9C6U1 | 2e-76 | MYB83_ARATH; Transcription factor MYB83 | ||||
| TrEMBL | A0A2G5DYB9 | 0.0 | A0A2G5DYB9_AQUCA; Uncharacterized protein | ||||
| STRING | Aquca_014_00829.1 | 0.0 | (Aquilegia coerulea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G12870.1 | 8e-69 | myb domain protein 46 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aqcoe2G014200.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



