 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aqcoe2G084900.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Ranunculales; Ranunculaceae; Thalictroideae; Aquilegia
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 366aa MW: 41753.5 Da PI: 8.6198 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 102.4 | 2.8e-32 | 78 | 131 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
prlrWtp+LH rFv+ave+LGG+e+AtPk++l++m++kgL+++hvkSHLQ++R+
Aqcoe2G084900.1.p 78 PRLRWTPDLHLRFVHAVERLGGQERATPKQVLQMMNIKGLSIAHVKSHLQMFRS 131
8****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 2.7E-29 | 74 | 132 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 11.783 | 74 | 134 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 3.76E-15 | 75 | 132 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 4.1E-23 | 78 | 133 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 8.1E-9 | 79 | 129 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 366 aa Download sequence Send to blast |
MSSMVIEATE EVRSKMKDSV SSDGNSITKS LENIEDKEDE VDQEQPMNGS SSSNSTVEEN 60 ERNKSSTGVR QYVRSKTPRL RWTPDLHLRF VHAVERLGGQ ERATPKQVLQ MMNIKGLSIA 120 HVKSHLQMFR SKKIDDQGQV INDRRHLMGS WDHNIHNPRQ LDHMLQGFTH RLASNYSDAS 180 WTTSHMSLAH SLCNGRENNV RDRLRYYASL AERNFAGSSS ERKTSLSTNF LVRNEPNSMT 240 TSRLHADQFQ LRYLQHSCQP LLEQRLRESS YISQSNARRE HMKSLNNADT TICRITQEER 300 NMTAKRKVED CDLDLNLSLN VRPGRSSDWT KGLGEEDTDI NLSLSLLPPS KKGKNSIDLN 360 NLQAE* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4r_A | 6e-17 | 79 | 133 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 6e-17 | 79 | 133 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 6e-17 | 79 | 133 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 6e-17 | 79 | 133 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00297 | DAP | Transfer from AT2G38300 | Download |
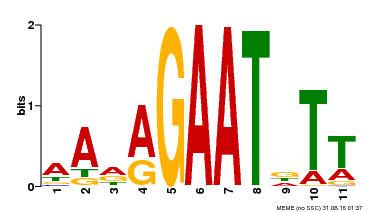
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019077022.1 | 1e-75 | PREDICTED: putative Myb family transcription factor At1g14600 | ||||
| TrEMBL | A0A2G5EUY4 | 0.0 | A0A2G5EUY4_AQUCA; Uncharacterized protein | ||||
| STRING | Aquca_004_00431.1 | 0.0 | (Aquilegia coerulea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G38300.1 | 4e-49 | G2-like family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aqcoe2G084900.1.p |



