 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aqcoe2G150600.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Ranunculales; Ranunculaceae; Thalictroideae; Aquilegia
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 558aa MW: 61956.2 Da PI: 6.1569 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 50.3 | 5.4e-16 | 59 | 105 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g WT++Ede l av+ + g++Wk+Ia+ ++ R++ qc +rwqk+l
Aqcoe2G150600.1.p 59 KGGWTPQEDETLRSAVEAFKGKCWKKIAEFFP-DRSEVQCLHRWQKVL 105
688*****************************.************986 PP
| |||||||
| 2 | Myb_DNA-binding | 61.8 | 1.4e-19 | 111 | 157 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEd++++++v+++G+ W+ Ia+ ++ gR +kqc++rw+++l
Aqcoe2G150600.1.p 111 KGPWTQEEDDKIIELVAKYGPTKWSVIAKSLP-GRIGKQCRERWHNHL 157
79******************************.*************97 PP
| |||||||
| 3 | Myb_DNA-binding | 52.2 | 1.4e-16 | 163 | 206 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
+++WT eE++ l +a +++G++ W+ Ia+ ++ gRt++++k++w++
Aqcoe2G150600.1.p 163 KDAWTLEEELSLMNAQREYGNK-WAEIAKVLP-GRTDNSIKNHWNS 206
679*******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 15.007 | 54 | 105 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.66E-14 | 55 | 112 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.4E-14 | 58 | 107 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 9.4E-15 | 59 | 105 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.1E-19 | 61 | 111 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.98E-12 | 62 | 105 | No hit | No description |
| PROSITE profile | PS51294 | 31.302 | 106 | 161 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.24E-31 | 108 | 204 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 6.3E-19 | 110 | 159 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 4.8E-19 | 111 | 157 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.1E-27 | 112 | 160 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 2.35E-16 | 113 | 157 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 8.0E-22 | 161 | 212 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 3.2E-15 | 162 | 210 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 21.49 | 162 | 212 | IPR017930 | Myb domain |
| Pfam | PF00249 | 5.5E-15 | 163 | 206 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.22E-11 | 165 | 208 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 558 aa Download sequence Send to blast |
MTELKIQQIC LVNKQSEETS SSSFSEESYS IALKSPEITS PVTTSPSHRR TSGPIRRAKG 60 GWTPQEDETL RSAVEAFKGK CWKKIAEFFP DRSEVQCLHR WQKVLNPELI KGPWTQEEDD 120 KIIELVAKYG PTKWSVIAKS LPGRIGKQCR ERWHNHLNPM IKKDAWTLEE ELSLMNAQRE 180 YGNKWAEIAK VLPGRTDNSI KNHWNSSLKK KLDFFVATGR LPPVPTTGPN GAKDTCKTTA 240 AGQISACSSK VSDSTDEKCV ESEDLCKAEG DGKDRLEDLA LECLTDTATG VALIGSSRSD 300 DGQCTRASII RCTESVSTPN PVDIENTGGD DSDNKLDGSP VHSTIRTFGS LYYEPPQFEN 360 IDIPLSSALF DTNCSLHRIY SANSVTSPSG YATPPCVEDS NFGLRNAESI LRNAAKSFPN 420 TPSILRKRKT EAYVTLPQDG ILKGNEMKVL DSSNGLKEKE ISDQDRFVRR SEGEKLYQNP 480 AFHGNDTIEY CDGENFNTAP PYRIWSKRTS VYKSLEKQLD FTLDTQNFES QAQSSLVIKG 540 NQPFTESYSQ TAKMGVA* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h88_C | 1e-68 | 58 | 212 | 5 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 1e-68 | 58 | 212 | 5 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00482 | DAP | Transfer from AT5G02320 | Download |
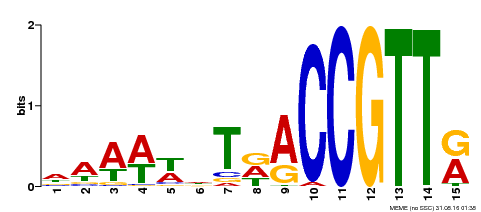
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010256935.1 | 0.0 | PREDICTED: myb-related protein B | ||||
| Refseq | XP_010256936.1 | 0.0 | PREDICTED: myb-related protein B | ||||
| TrEMBL | A0A2G5EX12 | 0.0 | A0A2G5EX12_AQUCA; Uncharacterized protein | ||||
| STRING | Aquca_003_00045.1 | 0.0 | (Aquilegia coerulea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G02320.2 | 1e-138 | myb domain protein 3r-5 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aqcoe2G150600.1.p |



