 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aqcoe2G263300.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Ranunculales; Ranunculaceae; Thalictroideae; Aquilegia
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 330aa MW: 37602 Da PI: 7.3222 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 58.5 | 1.6e-18 | 28 | 75 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg+WT eEd+ l++++ +G g+W++ ar+ g++Rt+k+c++rw++yl
Aqcoe2G263300.1.p 28 RGPWTVEEDLTLINYISNHGEGRWNSLARCAGLKRTGKSCRLRWLNYL 75
89********************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 51.2 | 2.8e-16 | 81 | 124 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
rg+ T eE++l+++++ ++G++ W++Ia++++ gRt++++k++w++
Aqcoe2G263300.1.p 81 RGNITLEEQLLILELHSRWGNR-WSKIAQHLP-GRTDNEIKNYWRT 124
7999******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 15.864 | 23 | 75 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.05E-30 | 25 | 122 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 4.5E-15 | 27 | 77 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.2E-16 | 28 | 75 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.1E-23 | 29 | 82 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 5.42E-11 | 30 | 75 | No hit | No description |
| PROSITE profile | PS51294 | 23.246 | 76 | 130 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.3E-14 | 80 | 128 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 5.7E-15 | 81 | 124 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 7.9E-23 | 83 | 129 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 6.96E-7 | 101 | 124 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009620 | Biological Process | response to fungus | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 330 aa Download sequence Send to blast |
MDINVRGCSS SSSATTQSDV EDSLDLRRGP WTVEEDLTLI NYISNHGEGR WNSLARCAGL 60 KRTGKSCRLR WLNYLRPDVR RGNITLEEQL LILELHSRWG NRWSKIAQHL PGRTDNEIKN 120 YWRTRVQKHA KQLKCDVNSK QFKDTMRYLW MPRLVERIQA AAATSSASGS NTTTTTTTSH 180 HHLYNNNNNS NSSYNNNNSS NNSSYNNQEL TTATQILPVG RGAFINSSFT PENSSTGASS 240 DSMASDMRDY YTFQANNNNY QNTQNAHMNY PDSQTNLSGY FNQELAYQGS DQHQQQQNNM 300 WFGGGEDIVE NSWNVEDLWF LQQQLMHGN* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h8a_C | 3e-25 | 25 | 122 | 24 | 120 | MYB TRANSFORMING PROTEIN |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in abiotic stress responses. Plays a regulatory role in tolerance to salt, cold, and drought stresses. Regulates positively the expression of genes involved in proline synthesis and transport, and genes involved in reactive oxygen species (ROS) scavenging such as peroxidase, superoxide dismutase and catalase during salt stress. Transactivates stress-related genes, including LEA3, RAB16A and DREB2A during salt stress. {ECO:0000269|PubMed:22301384}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00334 | DAP | Transfer from AT3G06490 | Download |
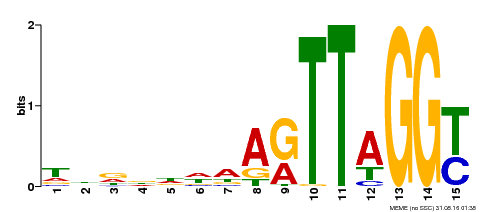
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by salt, cold and osmotic stresses, and abscisic acid (ABA). Down-regulated by salicylic acid (SA). {ECO:0000269|PubMed:22301384}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_028115735.1 | 1e-126 | transcription factor MYB108-like | ||||
| Swissprot | Q10MB4 | 5e-92 | MYB2_ORYSJ; Transcription factor MYB2 | ||||
| TrEMBL | A0A2G5EYT2 | 0.0 | A0A2G5EYT2_AQUCA; Uncharacterized protein | ||||
| STRING | Aquca_003_00440.1 | 0.0 | (Aquilegia coerulea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G49620.1 | 4e-85 | myb domain protein 78 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aqcoe2G263300.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



