 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aqcoe2G277000.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Ranunculales; Ranunculaceae; Thalictroideae; Aquilegia
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 913aa MW: 103558 Da PI: 6.7817 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 163.7 | 3.2e-51 | 30 | 146 | 2 | 117 |
CG-1 2 lke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenp 92
++e k+rwl+++ei+aiL n++ ++++ ++ + p+sg ++L++rkk+r+frkDG++wkkkkdgktv+E+he+LKvg+ e +++yYah+e+np
Aqcoe2G277000.1.p 30 MEEaKTRWLRPNEIHAILYNHNIFTIHVKPVHLPRSGLIVLFDRKKLRNFRKDGHNWKKKKDGKTVKEAHEHLKVGNEERIHVYYAHGEDNP 121
55669*************************************************************************************** PP
CG-1 93 tfqrrcywlLeeelekivlvhylev 117
f rrcywlL++++e++vlvhy+e+
Aqcoe2G277000.1.p 122 GFVRRCYWLLDKTYEHVVLVHYRET 146
***********************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 75.863 | 26 | 152 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 2.2E-71 | 29 | 147 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 1.8E-44 | 32 | 145 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 1.5E-6 | 362 | 454 | IPR013783 | Immunoglobulin-like fold |
| SuperFamily | SSF81296 | 2.94E-15 | 367 | 453 | IPR014756 | Immunoglobulin E-set |
| CDD | cd00204 | 2.58E-15 | 535 | 660 | No hit | No description |
| Pfam | PF12796 | 2.9E-7 | 548 | 629 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 2.02E-17 | 550 | 667 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 1.3E-17 | 558 | 665 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 18.324 | 559 | 664 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 1.9E-5 | 601 | 630 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 12.075 | 601 | 633 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 1000 | 640 | 670 | IPR002110 | Ankyrin repeat |
| SMART | SM00015 | 230 | 711 | 735 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 6.76 | 749 | 775 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 70 | 764 | 786 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 6.577 | 765 | 794 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.074 | 787 | 809 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.444 | 788 | 812 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.012 | 789 | 809 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 2.7 | 868 | 890 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.773 | 869 | 898 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.13 | 870 | 890 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 913 aa Download sequence Send to blast |
MDNNSASVRV AGAEIHGFHT LEDLDVAKMM EEAKTRWLRP NEIHAILYNH NIFTIHVKPV 60 HLPRSGLIVL FDRKKLRNFR KDGHNWKKKK DGKTVKEAHE HLKVGNEERI HVYYAHGEDN 120 PGFVRRCYWL LDKTYEHVVL VHYRETSELQ DSPVTPPNSS SASSYSDSSS RLVSEETDSG 180 ADRAYYMDPE TPFGGESTDR GERMIVQNHE VRLHEINTLE WEDLLVSNDP NNFNITSQDD 240 ISFLQQQNHS EMYDSHNNSL QPIAESGSLG VGLPEDGHLL NKGTNSGVQN QEFDMVTEHH 300 DDSAHVVQDG FGTQDSFGRW MNGIVTDSPG SLNNLPVGST ISSGHESNTS TVLDPYQFLT 360 QQQIFSITDI SPAWSFTAEE TKVIVVGYFH PAHSHFAETN LFCVFGDVCV PAEMIQVGVI 420 RCRALPHNPG IVNFYLSFDG HTPISQVMTF EYRASILENG LSPHEDNKWE EFQVQIRLSR 480 LLFSTTNSLN MLLSNISPND LKEAKKFATA TSSVDKDWAY LMKSIGKNEI SFPQAKNNLF 540 EIILKNKLQE WLLCRVVEGC QITMRDRQGQ GVLHLCAILG YTWAVRPYSH SGLSLDFRDA 600 SGWTALHWAA FYGREKMVAI LLSAGANPSL VSDPTSEFPG GCNAADLASK NGYEGLAAYL 660 AEKGLTEHFR LMSVCGNISG SLQSNSTDLV NPGNLSEEQL CQKDTLTAYR TAAEAASRIQ 720 SAFRENSFKL RKKAVEIATP ETEARDIIAA MKIQHAFRNY DTRKKIAAAG RIQYRFRTWK 780 IRKDFLNMRR QAIKIQAAFR ALQVRKHYHK ILWSVGVLEK GILRWRQKRK GFRGLQVEST 840 EVVDGEQKKE SDVEDDFFRI SRKQAEDRIE RSVVRVQALF RSFRAQQEYR RMKMAYDQVK 900 LEELLDTEVG FD* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 72 | 89 | RKKLRNFRKDGHNWKKKK |
| 2 | 73 | 88 | KKLRNFRKDGHNWKKK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator (PubMed:14581622). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in response to cold. Contributes together with CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:28351986, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
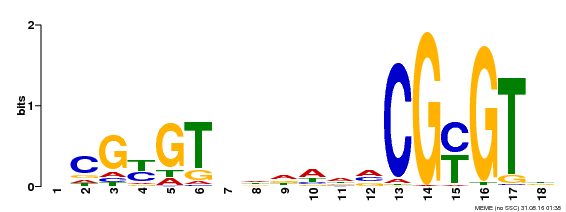
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, wounding, abscisic acid, H(2)O(2) and salicylic acid. {ECO:0000269|PubMed:12218065}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010259340.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 5 isoform X2 | ||||
| Swissprot | O23463 | 0.0 | CMTA5_ARATH; Calmodulin-binding transcription activator 5 | ||||
| TrEMBL | A0A2G5EZD7 | 0.0 | A0A2G5EZD7_AQUCA; Uncharacterized protein | ||||
| STRING | Aquca_003_00568.1 | 0.0 | (Aquilegia coerulea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aqcoe2G277000.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



