 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aqcoe2G277000.2.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Ranunculales; Ranunculaceae; Thalictroideae; Aquilegia
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 871aa MW: 98497.6 Da PI: 6.6474 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 131.7 | 2.6e-41 | 21 | 104 | 34 | 117 |
CG-1 34 pksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqrrcywlLeeelekivlvhylev 117
+ g ++L++rkk+r+frkDG++wkkkkdgktv+E+he+LKvg+ e +++yYah+e+np f rrcywlL++++e++vlvhy+e+
Aqcoe2G277000.2.p 21 LEGGLIVLFDRKKLRNFRKDGHNWKKKKDGKTVKEAHEHLKVGNEERIHVYYAHGEDNPGFVRRCYWLLDKTYEHVVLVHYRET 104
567999****************************************************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01076 | 4.2E-42 | 1 | 105 | IPR005559 | CG-1 DNA-binding domain |
| PROSITE profile | PS51437 | 58.588 | 1 | 110 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 4.9E-34 | 23 | 103 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 1.4E-6 | 320 | 412 | IPR013783 | Immunoglobulin-like fold |
| SuperFamily | SSF81296 | 2.76E-15 | 325 | 411 | IPR014756 | Immunoglobulin E-set |
| CDD | cd00204 | 1.33E-15 | 493 | 618 | No hit | No description |
| Pfam | PF12796 | 2.7E-7 | 506 | 587 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 1.87E-17 | 508 | 625 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 1.4E-17 | 516 | 623 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 18.324 | 517 | 622 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 1.9E-5 | 559 | 588 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 12.075 | 559 | 591 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 1000 | 598 | 628 | IPR002110 | Ankyrin repeat |
| SMART | SM00015 | 230 | 669 | 693 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 6.76 | 707 | 733 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 70 | 722 | 744 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 6.577 | 723 | 752 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.074 | 745 | 767 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.444 | 746 | 770 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.011 | 747 | 767 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 2.7 | 826 | 848 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.773 | 827 | 856 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.13 | 828 | 848 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 871 aa Download sequence Send to blast |
MDNNSASVRV AGAEIHGFHT LEGGLIVLFD RKKLRNFRKD GHNWKKKKDG KTVKEAHEHL 60 KVGNEERIHV YYAHGEDNPG FVRRCYWLLD KTYEHVVLVH YRETSELQDS PVTPPNSSSA 120 SSYSDSSSRL VSEETDSGAD RAYYMDPETP FGGESTDRGE RMIVQNHEVR LHEINTLEWE 180 DLLVSNDPNN FNITSQDDIS FLQQQNHSEM YDSHNNSLQP IAESGSLGVG LPEDGHLLNK 240 GTNSGVQNQE FDMVTEHHDD SAHVVQDGFG TQDSFGRWMN GIVTDSPGSL NNLPVGSTIS 300 SGHESNTSTV LDPYQFLTQQ QIFSITDISP AWSFTAEETK VIVVGYFHPA HSHFAETNLF 360 CVFGDVCVPA EMIQVGVIRC RALPHNPGIV NFYLSFDGHT PISQVMTFEY RASILENGLS 420 PHEDNKWEEF QVQIRLSRLL FSTTNSLNML LSNISPNDLK EAKKFATATS SVDKDWAYLM 480 KSIGKNEISF PQAKNNLFEI ILKNKLQEWL LCRVVEGCQI TMRDRQGQGV LHLCAILGYT 540 WAVRPYSHSG LSLDFRDASG WTALHWAAFY GREKMVAILL SAGANPSLVS DPTSEFPGGC 600 NAADLASKNG YEGLAAYLAE KGLTEHFRLM SVCGNISGSL QSNSTDLVNP GNLSEEQLCQ 660 KDTLTAYRTA AEAASRIQSA FRENSFKLRK KAVEIATPET EARDIIAAMK IQHAFRNYDT 720 RKKIAAAGRI QYRFRTWKIR KDFLNMRRQA IKIQAAFRAL QVRKHYHKIL WSVGVLEKGI 780 LRWRQKRKGF RGLQVESTEV VDGEQKKESD VEDDFFRISR KQAEDRIERS VVRVQALFRS 840 FRAQQEYRRM KMAYDQVKLE ELLDTEVGFD * |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 30 | 47 | RKKLRNFRKDGHNWKKKK |
| 2 | 31 | 46 | KKLRNFRKDGHNWKKK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator (PubMed:14581622). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in response to cold. Contributes together with CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:28351986, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
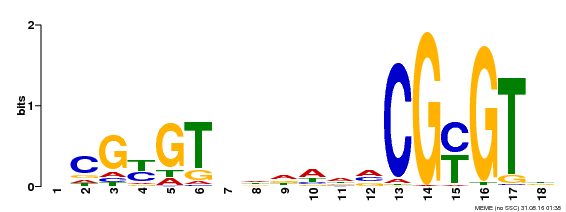
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, wounding, abscisic acid, H(2)O(2) and salicylic acid. {ECO:0000269|PubMed:12218065}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010259340.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 5 isoform X2 | ||||
| Swissprot | O23463 | 0.0 | CMTA5_ARATH; Calmodulin-binding transcription activator 5 | ||||
| TrEMBL | A0A2G5EZD7 | 0.0 | A0A2G5EZD7_AQUCA; Uncharacterized protein | ||||
| STRING | Aquca_003_00568.1 | 0.0 | (Aquilegia coerulea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aqcoe2G277000.2.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



