 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aqcoe2G282600.2.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Ranunculales; Ranunculaceae; Thalictroideae; Aquilegia
|
||||||||
| Family | ARR-B | ||||||||
| Protein Properties | Length: 637aa MW: 69424.2 Da PI: 6.0691 | ||||||||
| Description | ARR-B family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 79.4 | 4.4e-25 | 208 | 257 | 1 | 51 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQ 51
kpr++W+ eLH++Fv av+qL G++kA+Pk+ilelm+v+gLt+e+v+SHLQ
Aqcoe2G282600.2.p 208 KPRVVWSVELHQQFVAAVNQL-GIDKAVPKKILELMNVPGLTRENVASHLQ 257
79*******************.***************************** PP
| |||||||
| 2 | Response_reg | 73 | 1.2e-24 | 30 | 138 | 1 | 109 |
EEEESSSHHHHHHHHHHHHHTTCEEEEEESSHHHHHHHHHHHH..ESEEEEESSCTTSEHHHHHHHHHHHTTTSEEEEEESTTTHHHHHHHH CS
Response_reg 1 vlivdDeplvrellrqalekegyeevaeaddgeealellkekd..pDlillDiempgmdGlellkeireeepklpiivvtahgeeedaleal 90
vl+vdD+p+ +++l+++l++ y ev +++ + al +l+e++ +D++l D+ mp+mdG++ll++i e +lp+i+++ +++e + + +
Aqcoe2G282600.2.p 30 VLVVDDDPTCLKILEKMLRNCLY-EVITCSRAVVALSMLRERTysFDIVLSDVHMPDMDGFKLLEHIGLEM-DLPVIMMSVDDKKEVVMKGV 119
89*********************.99*************999999**********************6654.8******************* PP
HTTESEEEESS--HHHHHH CS
Response_reg 91 kaGakdflsKpfdpeelvk 109
Ga d+l Kp+ +e++++
Aqcoe2G282600.2.p 120 THGACDYLIKPVRIEAIRN 138
***************9987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:3.40.50.2300 | 1.1E-40 | 27 | 166 | No hit | No description |
| SuperFamily | SSF52172 | 3.56E-34 | 27 | 154 | IPR011006 | CheY-like superfamily |
| SMART | SM00448 | 2.6E-27 | 28 | 140 | IPR001789 | Signal transduction response regulator, receiver domain |
| PROSITE profile | PS50110 | 40.688 | 29 | 144 | IPR001789 | Signal transduction response regulator, receiver domain |
| Pfam | PF00072 | 5.1E-22 | 30 | 138 | IPR001789 | Signal transduction response regulator, receiver domain |
| CDD | cd00156 | 1.03E-25 | 31 | 143 | No hit | No description |
| SuperFamily | SSF46689 | 3.94E-16 | 205 | 257 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 9.395 | 205 | 270 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 4.2E-25 | 206 | 257 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.4E-21 | 208 | 257 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 4.5E-6 | 210 | 257 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009873 | Biological Process | ethylene-activated signaling pathway | ||||
| GO:0010082 | Biological Process | regulation of root meristem growth | ||||
| GO:0010119 | Biological Process | regulation of stomatal movement | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0071368 | Biological Process | cellular response to cytokinin stimulus | ||||
| GO:0080113 | Biological Process | regulation of seed growth | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 637 aa Download sequence Send to blast |
MLNMDSGGGG TSKKASGDGV SDQFPAGLRV LVVDDDPTCL KILEKMLRNC LYEVITCSRA 60 VVALSMLRER TYSFDIVLSD VHMPDMDGFK LLEHIGLEMD LPVIMMSVDD KKEVVMKGVT 120 HGACDYLIKP VRIEAIRNIW QHVVRKRRNE LKEFEQSGSY EDNDRNHKAS EDVDYASSVI 180 EGSWKGSKKR KEDEDDLEDR DDTSTMKKPR VVWSVELHQQ FVAAVNQLGI DKAVPKKILE 240 LMNVPGLTRE NVASHLQAGG LARSTVNTGI GMPHLDQRIP FSTEASKLRF GAGQQMTSSN 300 KQVDLYHGLP TNIEPKQLSH LHQSAQSSGS MGLQASEGTT GFMNLQTSLG THSMNQSGAV 360 LGSQSNSPLM MQMAQSRSRG QLLSDISSSH PSGTPLSIGQ QVFSNDMASR VLGKNITILN 420 GKATTYTPSF QASSMLDFQK KHTMDVPGNG FPVGITTGVS GLQSTGVFQD RGSLEVKGPR 480 SLPTGYVIFD EHPQHKSQEW GLQNVDLTFE ASQRPKPLQG NITFSPPVLA HQGLWSSQRG 540 GPNGNVSIVG NTLVSLSEGN QLGTSESFVQ RGNSLPMDSS FRVKAESMGD LNSDNIFAAK 600 TFDQDDLMSA ILKQQGGGQL ENDFNFDGYP IDDIPV* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 4e-19 | 204 | 257 | 1 | 54 | ARR10-B |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00054 | PBM | Transfer from AT4G16110 | Download |
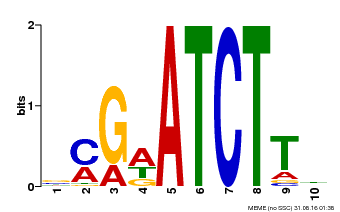
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010268090.1 | 0.0 | PREDICTED: two-component response regulator ARR2-like isoform X1 | ||||
| Refseq | XP_010268091.1 | 0.0 | PREDICTED: two-component response regulator ARR2-like isoform X2 | ||||
| TrEMBL | A0A2G5EZP7 | 0.0 | A0A2G5EZP7_AQUCA; Uncharacterized protein | ||||
| STRING | Aquca_003_00615.1 | 0.0 | (Aquilegia coerulea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16110.1 | 1e-124 | response regulator 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aqcoe2G282600.2.p |



