 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aqcoe2G351400.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Ranunculales; Ranunculaceae; Thalictroideae; Aquilegia
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 598aa MW: 68405.6 Da PI: 6.1543 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 43 | 1.2e-13 | 98 | 151 | 2 | 60 |
trihelix 2 WtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkr 60
W k+evlaL+++r+++++++++ W +vs+k++e g++rs+++Ckek+e+++k+
Aqcoe2G351400.1.p 98 WLKEEVLALLTIRSSLGNEFSDF-----IWGHVSRKLAELGYKRSAEKCKEKFEEVSKY 151
*********************98.....9****************************98 PP
| |||||||
| 2 | trihelix | 95.8 | 4e-30 | 447 | 532 | 1 | 86 |
trihelix 1 rWtkqevlaLiearremeerlrr.gklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdqle 86
rW+++ev Li++r ++++++ + ++ k+plWe++s+ m e g++rs+k+Ckekwen+nk+++k+k+++kkr s +s+tcpyf+ql+
Aqcoe2G351400.1.p 447 RWPRDEVNSLINLRCNLQSSVDDkESTKAPLWERISQGMLELGYKRSAKKCKEKWENINKYFRKTKDTNKKR-SLDSKTCPYFHQLN 532
8*********************735799*******************************************8.999*********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00717 | 0.21 | 94 | 151 | IPR001005 | SANT/Myb domain |
| Pfam | PF13837 | 8.9E-9 | 96 | 152 | No hit | No description |
| PROSITE profile | PS50090 | 6.516 | 97 | 149 | IPR017877 | Myb-like domain |
| SMART | SM00717 | 1.9 | 444 | 507 | IPR001005 | SANT/Myb domain |
| CDD | cd12203 | 1.23E-23 | 446 | 512 | No hit | No description |
| Pfam | PF13837 | 1.7E-19 | 446 | 533 | No hit | No description |
| PROSITE profile | PS50090 | 6.841 | 447 | 505 | IPR017877 | Myb-like domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0001158 | Molecular Function | enhancer sequence-specific DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 598 aa Download sequence Send to blast |
MFNGVPEQFH QFIASRSTLP LPLPLTLPLH VSPPPPTFSV HELQQQPSHQ VFQSHLLQPL 60 HRQTSTHESE EKEEAERVVG VNLEIEGMRS ASELMDPWLK EEVLALLTIR SSLGNEFSDF 120 IWGHVSRKLA ELGYKRSAEK CKEKFEEVSK YCNNNSYGNY YRSFNELEAL FEDQNPKSID 180 DDKLVRNTND DQEKMEVNME ENSANETLEN PIEENQTEEN QYVVKKPKKK KKKYHQIELL 240 KGFSEFVNKI MVQQEELHKK LLEDMERREE ERVAREEAWK KKEMDRFFKE MEIRAQEQAI 300 ACDRESKIVE FLNQFTSNIS QTQGLVVQKD NQVFKTPMGK STNFPTTSPS INVLPENPSS 360 SRISAKPNAP SSSAMILSPQ NKISSPTQNN PQMPNSSNLD PSSHHNPNPV NTQTNTNIPP 420 SSSTQLLPLN PKPINSASTD REDYGKRWPR DEVNSLINLR CNLQSSVDDK ESTKAPLWER 480 ISQGMLELGY KRSAKKCKEK WENINKYFRK TKDTNKKRSL DSKTCPYFHQ LNTLYNQGRL 540 TMPSEVPENR SVATPPPETG VGSGASHQDA GLSSEAIVHV NEEGEKNMVQ VPLEFEY* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 224 | 232 | KKPKKKKKK |
| 2 | 225 | 232 | KPKKKKKK |
| 3 | 490 | 498 | KRSAKKCKE |
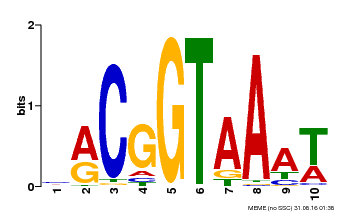
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00011 | PBM | Transfer from AT5G28300 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP009768 | 5e-37 | AP009768.1 Lotus japonicus genomic DNA, clone: LjT40E15, TM1928, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_026456984.1 | 0.0 | trihelix transcription factor GTL2-like | ||||
| TrEMBL | A0A2G5CK61 | 0.0 | A0A2G5CK61_AQUCA; Uncharacterized protein | ||||
| STRING | Aquca_049_00159.1 | 0.0 | (Aquilegia coerulea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G28300.1 | 9e-54 | Trihelix family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aqcoe2G351400.1.p |



