 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aqcoe2G375000.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Ranunculales; Ranunculaceae; Thalictroideae; Aquilegia
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 1140aa MW: 127900 Da PI: 4.8141 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 42.8 | 1.2e-13 | 8 | 53 | 2 | 48 |
SSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 2 grWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
g W Ede+l+ av ++G+++W++I++ + ++++kqck rw+ +l
Aqcoe2G375000.1.p 8 GVWKNTEDEILKAAVMKYGKNQWARISSLLV-RKSAKQCKARWYEWL 53
78*****************************.************986 PP
| |||||||
| 2 | Myb_DNA-binding | 32.6 | 1.9e-10 | 60 | 103 | 2 | 48 |
SSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 2 grWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
WT eEde+l+++ k++++ W+tIa +g Rt+ qc +r+ k+l
Aqcoe2G375000.1.p 60 TEWTREEDEKLLHLAKLMPTQ-WRTIAPIVG--RTPSQCLERYEKLL 103
68*****************99.********8..**********9986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 22.008 | 2 | 57 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 6.6E-18 | 5 | 56 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 7.0E-15 | 6 | 55 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 5.33E-12 | 10 | 53 | No hit | No description |
| Pfam | PF13921 | 5.7E-13 | 10 | 70 | No hit | No description |
| SuperFamily | SSF46689 | 8.99E-21 | 33 | 107 | IPR009057 | Homeodomain-like |
| CDD | cd11659 | 1.10E-30 | 55 | 106 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 3.8E-14 | 57 | 104 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 16.443 | 58 | 107 | IPR017930 | Myb domain |
| SMART | SM00717 | 1.7E-12 | 58 | 105 | IPR001005 | SANT/Myb domain |
| Pfam | PF11831 | 8.2E-56 | 426 | 672 | IPR021786 | Pre-mRNA splicing factor component Cdc5p/Cef1 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009870 | Biological Process | defense response signaling pathway, resistance gene-dependent | ||||
| GO:0010204 | Biological Process | defense response signaling pathway, resistance gene-independent | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0009507 | Cellular Component | chloroplast | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1140 aa Download sequence Send to blast |
MRIMIKGGVW KNTEDEILKA AVMKYGKNQW ARISSLLVRK SAKQCKARWY EWLDPSIKKT 60 EWTREEDEKL LHLAKLMPTQ WRTIAPIVGR TPSQCLERYE KLLDAACVKD ENYEPNDDPR 120 KLRPGEIDPN PESKPARPDP VDMDEDEKEM LSEARARLAN TRGKKAKRKA REKQLEEARR 180 LASLQKRREL KAAGIDTRQR KRKRKGIDYN AEIPFEKKPP AGFFDVADEE THVEQPKFPT 240 TIEELEGKRR VDVEAQLRKQ DIARNKIAQR QDAPSSILQV NKLNDPESVR KRSKLMLPAP 300 QITDHELEEI AKMGYASDIV SGSEHIGEGS GATRALLANY AQTPQQGMTP FRTPQRTPGG 360 KADAIMMEAE NLARLRESQT PLLGGDNPEL HPSDFSGVTP KKREIQTPHP MSTPSTPGAM 420 GLTPRSGMTP SAGLTPRIGM TPSRDGYSFG MTPKGTPIRD ELRINISEDM DVHDGAKHEL 480 RSQAELRRDL RSGLTSLPQP KNEYQIVVQP LPEENDEPED KVEEDMSDKI AREKAEEEAK 540 QQELLKKRSK VLQRELPRPP PASVELIKNS LMKSDEDKSS FVPPTSFEQA DEMIRKELLS 600 LLEHDNAKYP LEEKVGKEKK KGGKRAPSGK STTFVPEIEE FANDILKEAD SLINEEIEYL 660 RVAMGHGNES LDEFVKAHDA CQEDLMYFPT RYAYGLSSVA GNMEKLTALQ HEFENVKNRM 720 DDETKKVQRL EQKIKLLTHG YQTRGGKLWS QIEATFKQMN TAATELECFQ ALQKQEHYAA 780 SHRINVLFEE INRQKELEEN LQKRYGDLVA EKERIQRLLE EHKRQAQIQE EIDSQNCALE 840 ESKLQAQIQE EIAEKNRAFE LSISAADQVD NVQEQVTDAI PKDPGTDMDV DSHLDSTSLE 900 ANLTKDDMVV GKAMLVQDSC CKGPNSSDQP LTADNDDTSG VIVQILNVPV QEVALNGGPA 960 PTLLGTEEMH MTSDSVMDQM PPGIGDSLKI SQEEINVMSE NSHMESKILV EEQLVVMEAG 1020 TQDGIVMEAE ASLRHSSMDS LKVSQEELNA TSENSNMESK ILVEEQLVVT GAGMQDGIVT 1080 KSEASLEDSL DAGRENITIS SSEKRIVVNL DGEADENLKC GVDAVSNNET MPVLNEDLS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5mqf_L | 0.0 | 2 | 816 | 3 | 799 | Cell division cycle 5-like protein |
| 5xjc_L | 0.0 | 2 | 816 | 3 | 799 | Cell division cycle 5-like protein |
| 5yzg_L | 0.0 | 2 | 816 | 3 | 799 | Cell division cycle 5-like protein |
| 5z56_L | 0.0 | 2 | 816 | 3 | 799 | Cell division cycle 5-like protein |
| 5z57_L | 0.0 | 2 | 816 | 3 | 799 | Cell division cycle 5-like protein |
| 5z58_L | 0.0 | 2 | 816 | 3 | 799 | Cell division cycle 5-like protein |
| 6ff4_L | 0.0 | 2 | 816 | 3 | 799 | Cell division cycle 5-like protein |
| 6ff7_L | 0.0 | 2 | 816 | 3 | 799 | Cell division cycle 5-like protein |
| 6icz_L | 0.0 | 2 | 816 | 3 | 799 | Cell division cycle 5-like protein |
| 6id0_L | 0.0 | 2 | 816 | 3 | 799 | Cell division cycle 5-like protein |
| 6id1_L | 0.0 | 2 | 816 | 3 | 799 | Cell division cycle 5-like protein |
| 6qdv_O | 0.0 | 2 | 816 | 3 | 799 | Cell division cycle 5-like protein |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 185 | 204 | KRRELKAAGIDTRQRKRKRK |
| 2 | 199 | 204 | RKRKRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
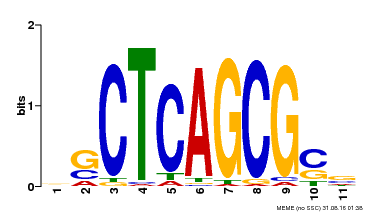
| UniProt | Component of the MAC complex that probably regulates defense responses through transcriptional control and thereby is essential for plant innate immunity. Possesses a sequence specific DNA sequence 'CTCAGCG' binding activity. Involved in mRNA splicing and cell cycle control. May also play a role in the response to DNA damage. {ECO:0000250|UniProtKB:Q99459, ECO:0000269|PubMed:17298883, ECO:0000269|PubMed:17575050, ECO:0000269|PubMed:8917598}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00028 | SELEX | Transfer from AT1G09770 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010246794.1 | 0.0 | PREDICTED: cell division cycle 5-like protein | ||||
| Swissprot | P92948 | 0.0 | CDC5L_ARATH; Cell division cycle 5-like protein | ||||
| TrEMBL | A0A2G5CYR2 | 0.0 | A0A2G5CYR2_AQUCA; Uncharacterized protein | ||||
| STRING | Aquca_034_00352.1 | 0.0 | (Aquilegia coerulea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09770.1 | 0.0 | cell division cycle 5 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aqcoe2G375000.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



