 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aqcoe2G433200.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Ranunculales; Ranunculaceae; Thalictroideae; Aquilegia
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 496aa MW: 53078.4 Da PI: 6.1926 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 33.8 | 7.1e-11 | 255 | 313 | 5 | 63 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklksev 63
kr++r NR +A rs +RK+ +i eLe+kv++L++e ++L +l+ l+ + l+ e+
Aqcoe2G433200.1.p 255 KRAKRIWANRQSAARSKERKLRYISELERKVQTLQTEATTLNTQLNVLQRDTSGLTVEN 313
9*********************************************9998888777766 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 5.9E-16 | 251 | 315 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 10.749 | 253 | 316 | IPR004827 | Basic-leucine zipper domain |
| Gene3D | G3DSA:1.20.5.170 | 2.4E-10 | 255 | 309 | No hit | No description |
| Pfam | PF00170 | 2.6E-9 | 255 | 309 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 7.74E-11 | 255 | 306 | No hit | No description |
| CDD | cd14703 | 3.48E-22 | 256 | 305 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 496 aa Download sequence Send to blast |
MDFDKPSFSS SSSSQHLHGG GVGGGGGLAP SPPLPPLSST TTRYFSSLGE SMAATATLES 60 GNNLINIENY TSDGNGNGNG NGIGNGNGNG NRFSYDISRM SDLPTKGLFG HRRAHSEILS 120 LPDDISFDSD LGVVGSGSAA NADGPSFSDE TEEDLFSMFM DMEKFGSGST ATSSGEPSSS 180 VPPPPPLGSG VQMDNSIMGF TDKPRIRHHH SQSMDGSTSI KSELLISGSD GPSFTENKKA 240 ISSSKLADLA LVDPKRAKRI WANRQSAARS KERKLRYISE LERKVQTLQT EATTLNTQLN 300 VLQRDTSGLT VENNELKLRL STVEQQVNLQ DALNNALRDE LQRLKVATGQ AITGVGSLMN 360 FGSTHRSSQP FYPNNQTMHG VLAAHQFQQL QIQSQQSLQL QQQQQQQLQQ QQTGDMKLKA 420 SLSSASRRDN SYDNNPSQRS GNSQPFSCPP ISAAPDSLST VPPVSTAAAT TSPAPSWGLE 480 TQRFSSLSEM QREFI* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Putative transcription factor with an activatory role. | |||||
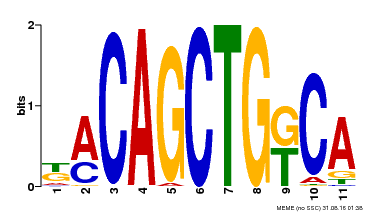
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00126 | DAP | Transfer from AT1G06070 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC253894 | 0.0 | AC253894.1 Aquilegia coerulea clone COL02-M12, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010268265.1 | 1e-136 | PREDICTED: probable transcription factor PosF21 | ||||
| Refseq | XP_010268266.1 | 1e-136 | PREDICTED: probable transcription factor PosF21 | ||||
| Refseq | XP_010268267.1 | 1e-136 | PREDICTED: probable transcription factor PosF21 | ||||
| Swissprot | Q04088 | 1e-123 | POF21_ARATH; Probable transcription factor PosF21 | ||||
| TrEMBL | A0A2G5CEK1 | 0.0 | A0A2G5CEK1_AQUCA; Uncharacterized protein | ||||
| STRING | Aquca_058_00063.1 | 0.0 | (Aquilegia coerulea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G06070.1 | 1e-121 | bZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aqcoe2G433200.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



