 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aqcoe3G360300.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Ranunculales; Ranunculaceae; Thalictroideae; Aquilegia
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 311aa MW: 34967.4 Da PI: 6.4068 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 99.9 | 1.6e-31 | 171 | 228 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
+Dgy+WrKYGqK vk+s++prsYYrCt ++C+vkk+vers edp++v++tYeg+H h+
Aqcoe3G360300.1.p 171 EDGYRWRKYGQKAVKNSPYPRSYYRCTNSKCTVKKRVERSCEDPTIVITTYEGQHCHH 228
8******************************************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 1.7E-32 | 155 | 229 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 2.22E-27 | 162 | 228 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 29.516 | 165 | 230 | IPR003657 | WRKY domain |
| SMART | SM00774 | 7.0E-36 | 170 | 229 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 2.1E-24 | 171 | 228 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 311 aa Download sequence Send to blast |
MDDDKKRKLE SEMISDSSSW VDSSYFFENE ERQRSILNEF GWNLPQQQQQ TETNQETTSF 60 MEFDDNQIEL EQQQQQRSDL AGSFLLPTEN HSTSSTTPTV IATTTTVEKS GDASTSNPSV 120 SSSSSDELPE KSTGSDGKPP EKPSKGRKKG QKRIRQQRFA FMTKSDVDHL EDGYRWRKYG 180 QKAVKNSPYP RSYYRCTNSK CTVKKRVERS CEDPTIVITT YEGQHCHHTV SFPRGSVFSH 240 EGSFGGGLNT TSQIYAPAMQ YTVSGPLSVT QSHQPQLEVR PAQMQPQQTL QNPSDEGLLG 300 DIVPPGMRNR * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 1e-25 | 160 | 227 | 7 | 74 | Probable WRKY transcription factor 4 |
| 2lex_A | 1e-25 | 160 | 227 | 7 | 74 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00067 | PBM | Transfer from AT1G69310 | Download |
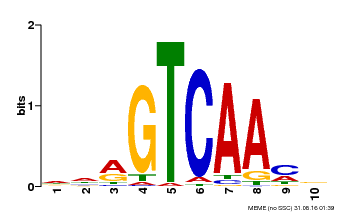
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021299838.1 | 1e-102 | probable WRKY transcription factor 57 | ||||
| Refseq | XP_021299839.1 | 1e-102 | probable WRKY transcription factor 57 | ||||
| Swissprot | Q9C983 | 1e-66 | WRK57_ARATH; Probable WRKY transcription factor 57 | ||||
| TrEMBL | A0A2G5F543 | 0.0 | A0A2G5F543_AQUCA; Uncharacterized protein | ||||
| STRING | Aquca_002_00789.1 | 0.0 | (Aquilegia coerulea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G69310.2 | 1e-54 | WRKY DNA-binding protein 57 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aqcoe3G360300.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



