 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aqcoe3G398400.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Ranunculales; Ranunculaceae; Thalictroideae; Aquilegia
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 331aa MW: 37374.1 Da PI: 8.7159 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 97 | 1.3e-30 | 159 | 214 | 1 | 56 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRla 56
k+r+ W++eLH++F++a++qLGGs +AtPk+i+elmkv+gLt+++vkSHLQkYRl+
Aqcoe3G398400.1.p 159 KARRSWSNELHKKFLNALQQLGGSYVATPKQIRELMKVDGLTNDEVKSHLQKYRLH 214
689***************************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 11.96 | 156 | 216 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.87E-17 | 157 | 217 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 4.8E-27 | 157 | 217 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.5E-26 | 159 | 214 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 1.1E-7 | 161 | 212 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0055062 | Biological Process | phosphate ion homeostasis | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 331 aa Download sequence Send to blast |
MDFIKKMKGI RCEDYMEALE EERHKIQVFH RELPLCLELV TQAIDICKKQ VGIKHECEDV 60 ELISSTETRK LYSPKEKKVD WLKSAQLWNH DFSDQKDKPS QNTIGIEPNR LKGAFRPFQR 120 DEGHRASTAT STSTTAETVG VSPVNRAEEK QRQVESHKKA RRSWSNELHK KFLNALQQLG 180 GSYVATPKQI RELMKVDGLT NDEVKSHLQK YRLHTRRTSA AIHIKDNLHS TQLILVEGLL 240 VQPEESFQDR PTTGDAAQLV TNGVYAPVAA RPQSPLLTSQ RQHKQPSTVH PESKIRGSQA 300 QHCMEDEGTH SDHQSSSSST QSTTVSVSLV * |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in phosphate homeostasis. Involved in the regulation of the developmental response of lateral roots, acquisition and/or mobilization of phosphate and expression of a subset of genes involved in phosphate sensing and signaling pathway. Is a target of the transcription factor PHR1. {ECO:0000269|PubMed:27016098}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00218 | DAP | Transfer from AT1G68670 | Download |
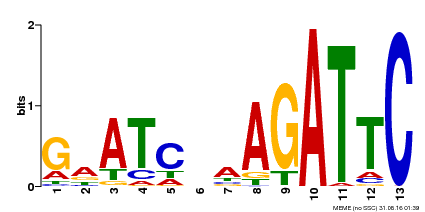
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced under phosphate deprivation conditions. {ECO:0000269|PubMed:27016098}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003631224.1 | 1e-79 | PREDICTED: myb family transcription factor EFM | ||||
| Swissprot | Q8VZS3 | 1e-68 | HHO2_ARATH; Transcription factor HHO2 | ||||
| TrEMBL | A0A2G5F6C0 | 1e-138 | A0A2G5F6C0_AQUCA; Uncharacterized protein | ||||
| STRING | Aquca_002_01118.1 | 1e-139 | (Aquilegia coerulea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G68670.1 | 2e-68 | G2-like family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aqcoe3G398400.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



