 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aqcoe3G427600.3.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Ranunculales; Ranunculaceae; Thalictroideae; Aquilegia
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 549aa MW: 59102.8 Da PI: 8.6182 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 16.3 | 2.8e-05 | 6 | 28 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
+ C+ C+k F r nL+ H r H
Aqcoe3G427600.3.p 6 FICEVCNKGFQREQNLQLHRRGH 28
89*******************88 PP
| |||||||
| 2 | zf-C2H2 | 12.6 | 0.00041 | 82 | 104 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
+kC +C+k++ +s+ k H +t+
Aqcoe3G427600.3.p 82 WKCDKCSKRYAVQSDWKAHSKTC 104
58*****************9998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:3.30.160.60 | 6.0E-6 | 5 | 28 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 3.61E-7 | 5 | 28 | No hit | No description |
| Pfam | PF12171 | 2.7E-5 | 6 | 28 | IPR022755 | Zinc finger, double-stranded RNA binding |
| PROSITE profile | PS50157 | 10.99 | 6 | 28 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0052 | 6 | 28 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 8 | 28 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 2.7E-4 | 70 | 103 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 3.61E-7 | 77 | 102 | No hit | No description |
| SMART | SM00355 | 140 | 82 | 102 | IPR015880 | Zinc finger, C2H2-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 549 aa Download sequence Send to blast |
MATNRFICEV CNKGFQREQN LQLHRRGHNL PWKLKQKTTK EVKRKVYLCP EPTCVHHDPS 60 RALGDLTGIK KHFCRKHGEK KWKCDKCSKR YAVQSDWKAH SKTCGTREYR CDCGTLFSRR 120 DSFITHRAFC DALAQESARL PTGLNTMGSH LYGGSNMGLN LSQVGSQMSS LQDQNHTSND 180 ILRLGGAGAQ YDHLISPSNS SFRPPQPSPA SAFYLPSGSN QDYHEDSQSH HQLLQNKNFH 240 GLMQLPELQS NANNSSPAAA AAAANLFNLS FFSNSSTTSN MNNSSNANNN NSNNHLSSSG 300 LLMPDQFNSG NAGGEGTTLF SGNIMGDHMG NNVSSLYNTS VQSDNVLPQM SATALLQKAA 360 QMGSSTSGNS STLLRGFGSS SSSNARSDRP HMSANFRGSF SSGGGGGGEN LRSQMENDNH 420 LQDIMNNFAN GGSQIFGSGS NSFGASGGHG QETSYAGFNS NRLSLDQQHG NSNMSNMDEA 480 KMHQNQLSAS LGGSDRLTRD FLGVGGMVRS VSGGISQREQ HHGLDISSLD SEIKSATENR 540 SFGSGSLQ* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5b3h_C | 2e-33 | 78 | 142 | 3 | 67 | Zinc finger protein JACKDAW |
| 5b3h_F | 2e-33 | 78 | 142 | 3 | 67 | Zinc finger protein JACKDAW |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor acting as a positive regulator of the starch synthase SS4. Controls chloroplast development and starch granule formation (PubMed:22898356). Binds DNA via its zinc fingers (PubMed:24821766). Recognizes and binds to SCL3 promoter sequence 5'-AGACAA-3' to promotes its expression when in complex with RGA (PubMed:24821766). {ECO:0000269|PubMed:22898356, ECO:0000269|PubMed:24821766}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00255 | DAP | Transfer from AT2G02070 | Download |
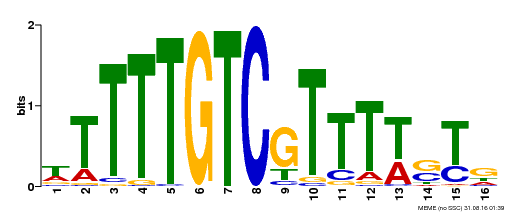
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC208494 | 1e-122 | AC208494.1 Populus trichocarpa clone JGIACSB265-L05, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010270755.1 | 0.0 | PREDICTED: protein indeterminate-domain 5, chloroplastic-like | ||||
| Refseq | XP_010270757.1 | 0.0 | PREDICTED: protein indeterminate-domain 5, chloroplastic-like | ||||
| Swissprot | Q9ZUL3 | 1e-119 | IDD5_ARATH; Protein indeterminate-domain 5, chloroplastic | ||||
| TrEMBL | A0A2G5F7R0 | 0.0 | A0A2G5F7R0_AQUCA; Uncharacterized protein | ||||
| STRING | Aquca_002_01389.1 | 0.0 | (Aquilegia coerulea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G02070.1 | 1e-102 | indeterminate(ID)-domain 5 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aqcoe3G427600.3.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



