 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aqcoe4G027100.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Ranunculales; Ranunculaceae; Thalictroideae; Aquilegia
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 329aa MW: 34779.8 Da PI: 8.0481 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 203.9 | 4.8e-63 | 2 | 146 | 1 | 149 |
DUF822 1 ggsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeaeaagssasaspes 92
++++r ptwkErEnnkrRERrRRaiaaki+aGLR++Gnyklpk++DnneVlkALc+eAGw+ve+DGttyrkg+kp e ++++g s ++sp+s
Aqcoe4G027100.1.p 2 TSGTRLPTWKERENNKRRERRRRAIAAKIFAGLRMYGNYKLPKHCDNNEVLKALCNEAGWTVEEDGTTYRKGCKPAEPMDMGGGSGTVSPCS 93
5899**************************************************************************************** PP
DUF822 93 slqsslkssalaspvesysaspksssfpspssldsislasa....asllpvlsvlslvsss 149
s+ +sp+ sy+ sp sssf sp+s+ +++ ++ sl+p+l++ls++sss
Aqcoe4G027100.1.p 94 SYR--------PSPCISYNVSPGSSSFASPVSSPYAPNGKStadgISLIPWLKNLSSASSS 146
*99........********************9998887766778899********997776 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 6.9E-64 | 3 | 141 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 329 aa Download sequence Send to blast |
MTSGTRLPTW KERENNKRRE RRRRAIAAKI FAGLRMYGNY KLPKHCDNNE VLKALCNEAG 60 WTVEEDGTTY RKGCKPAEPM DMGGGSGTVS PCSSYRPSPC ISYNVSPGSS SFASPVSSPY 120 APNGKSTADG ISLIPWLKNL SSASSSVSSA KLPHLYIPSG SISAPVTPPL SSPTARTPRF 180 NPDWEDTTAG PAWGGHRYSF LPSSTPPSPG RQVLPDNGWL AGIRLPTGGP SSPTFSLVSS 240 NPFGFKEGAS TGGGGSRMWT PGQSGTCSPA MPGAMDHTAD VPMSEGVSDE FAFESNTTGL 300 VKAWEGEIIH EECGSDELEL TLGTARTR* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5zd4_A | 2e-23 | 6 | 86 | 372 | 451 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_B | 2e-23 | 6 | 86 | 372 | 451 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_C | 2e-23 | 6 | 86 | 372 | 451 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_D | 2e-23 | 6 | 86 | 372 | 451 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00248 | DAP | Transfer from AT1G78700 | Download |
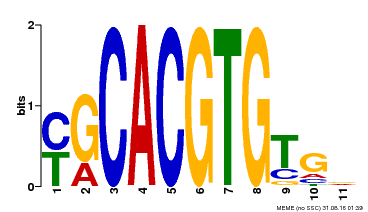
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010242162.1 | 1e-160 | PREDICTED: BES1/BZR1 homolog protein 4 | ||||
| Swissprot | Q9ZV88 | 1e-134 | BEH4_ARATH; BES1/BZR1 homolog protein 4 | ||||
| TrEMBL | A0A2G5CRJ2 | 0.0 | A0A2G5CRJ2_AQUCA; Uncharacterized protein | ||||
| STRING | Aquca_039_00027.1 | 0.0 | (Aquilegia coerulea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G78700.1 | 1e-124 | BES1/BZR1 homolog 4 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aqcoe4G027100.1.p |



