 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aqcoe5G151400.12.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Ranunculales; Ranunculaceae; Thalictroideae; Aquilegia
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 464aa MW: 50418.3 Da PI: 10.189 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 42.8 | 1.1e-13 | 407 | 458 | 5 | 56 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkev 56
+r+rr++kNRe+A rsR+RK+a++ eLe + ++L ++N++L+k+ e+ ++
Aqcoe5G151400.12.p 407 RRQRRMIKNRESAARSRARKQAYTMELEAELAKLREQNEELQKKQAEIMEMQ 458
79****************************************9999998874 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.20.1000.10 | 4.1E-4 | 241 | 291 | No hit | No description |
| SMART | SM00338 | 1.2E-9 | 403 | 462 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 11.346 | 405 | 463 | IPR004827 | Basic-leucine zipper domain |
| Gene3D | G3DSA:1.20.5.170 | 2.2E-13 | 407 | 456 | No hit | No description |
| SuperFamily | SSF57959 | 7.88E-10 | 407 | 455 | No hit | No description |
| CDD | cd14707 | 1.26E-26 | 407 | 461 | No hit | No description |
| Pfam | PF00170 | 1.8E-11 | 407 | 460 | IPR004827 | Basic-leucine zipper domain |
| PROSITE pattern | PS00036 | 0 | 410 | 425 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009738 | Biological Process | abscisic acid-activated signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 464 aa Download sequence Send to blast |
MGSQMNFRNI GDMSQQVDGS GGSGRPPGPP GNMSLARQTS IYSLTFEEFQ NTMGGFGKDF 60 GSMNMDEFLK NVWTAEETQV MGSTLGVPEG GVPLGGNLQR QGSLTLPRTL SQKTVDEVWR 120 DLFKENGGGV GKDGSVNGVA NMPQRQQTLG EMTLEEFLVR AGVVREEAQA VAQQNNGGFY 180 NELLPNNNNP GLSLGFGQPS QNNGIMANLI TGANNNNIAS NQSPNLAISI NGVRSPFVQQ 240 QQQQQQQQQQ QRQLQLQQQQ QLQQQQQQQK QQQQQKQQQQ QQQQQQQQQR QQQLFPKQPA 300 VSYASPLHLT NAQLASPGTR GVMMGMTNPT MNGNLVQGGG LQAGVLGTAG LGAGAVTVAS 360 GSPANQLSDG LGMSTGDMSS LSPVPYAFNG GLRGRRSGGT VEKVVERRQR RMIKNRESAA 420 RSRARKQAYT MELEAELAKL REQNEELQKK QAEIMEMQKL KVT* |
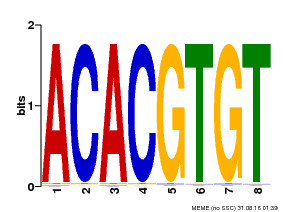
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00038 | PBM | Transfer from AT4G34000 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010269648.1 | 1e-149 | PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 5 isoform X1 | ||||
| Refseq | XP_010269652.1 | 1e-149 | PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 5 isoform X1 | ||||
| Refseq | XP_010269660.1 | 1e-149 | PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 5 isoform X1 | ||||
| Refseq | XP_019054745.1 | 1e-149 | PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 5 isoform X1 | ||||
| Refseq | XP_028806847.1 | 1e-149 | bZIP transcription factor TRAB1-like | ||||
| TrEMBL | A0A2G5EL64 | 0.0 | A0A2G5EL64_AQUCA; Uncharacterized protein | ||||
| STRING | Aquca_007_00660.1 | 0.0 | (Aquilegia coerulea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G19290.2 | 2e-46 | ABRE binding factor 4 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aqcoe5G151400.12.p |



