 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aqcoe5G197500.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Ranunculales; Ranunculaceae; Thalictroideae; Aquilegia
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 388aa MW: 41373.5 Da PI: 5.0477 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 71 | 1.8e-22 | 278 | 340 | 1 | 63 |
XXXXCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 1 ekelkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklksev 63
e+elkr+rrk +NRe+ArrsR+RK+ae+eeL+ kv++L+ eN+ L+kele+l +e +kl++e+
Aqcoe5G197500.1.p 278 ERELKRQRRKLSNRESARRSRLRKQAECEELQVKVDTLTDENDNLRKELERLAEERQKLTNEN 340
89**********************************************************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF07777 | 7.8E-27 | 1 | 92 | IPR012900 | G-box binding protein, multifunctional mosaic region |
| Pfam | PF16596 | 9.0E-12 | 113 | 243 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 3.0E-17 | 273 | 337 | No hit | No description |
| SMART | SM00338 | 2.6E-21 | 278 | 342 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 6.4E-20 | 278 | 340 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 13.633 | 280 | 343 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 2.31E-11 | 281 | 337 | No hit | No description |
| CDD | cd14702 | 5.65E-23 | 283 | 332 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 285 | 300 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010310 | Biological Process | regulation of hydrogen peroxide metabolic process | ||||
| GO:0010629 | Biological Process | negative regulation of gene expression | ||||
| GO:0090342 | Biological Process | regulation of cell aging | ||||
| GO:0005737 | Cellular Component | cytoplasm | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 388 aa Download sequence Send to blast |
MGSAEESTPA KPSRPSASSQ ETPPTPLYPD WSTPMQAYYG AGATQPPFFP TNVASPPPYP 60 YMWGGQMVSP YGTPIPYPAI YPHGGLYPHP NLATAQGAAM PTTQMEENNS PVKKIMSSGN 120 IGVVGGKLKE SGKAASGSRN DGVSRSAESG SEGSSDASDE FNHKDDSENK NKSFDQMLAD 180 GANAQNTSAH HSSAAVGSSL NGNGEPSANF PVPLPGNPVG DIAATNLNIG MDLWNASPVG 240 SVPLRARSNA SGVVPAVAPV KRDGHEGIVP EHLWGQDERE LKRQRRKLSN RESARRSRLR 300 KQAECEELQV KVDTLTDEND NLRKELERLA EERQKLTNEN ASLESELSQL YGEEAISTLK 360 GKNANMSVQS VNGFEQDTLM GNNSLSE* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 280 | 287 | LKRQRRKL |
| 2 | 294 | 300 | RRSRLRK |
| 3 | 294 | 301 | RRSRLRKQ |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00039 | PBM | Transfer from AT4G36730 | Download |
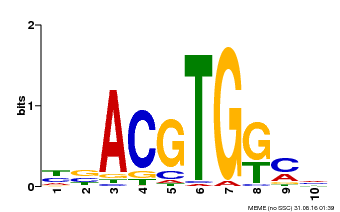
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010244323.1 | 1e-150 | PREDICTED: G-box-binding factor 1-like isoform X2 | ||||
| TrEMBL | A0A2G5EIR3 | 0.0 | A0A2G5EIR3_AQUCA; Uncharacterized protein | ||||
| STRING | Aquca_007_00161.1 | 0.0 | (Aquilegia coerulea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G36730.1 | 2e-70 | G-box binding factor 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aqcoe5G197500.1.p |



