 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aqcoe5G214000.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Ranunculales; Ranunculaceae; Thalictroideae; Aquilegia
|
||||||||
| Family | EIL | ||||||||
| Protein Properties | Length: 609aa MW: 67841.4 Da PI: 5.366 | ||||||||
| Description | EIL family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | EIN3 | 427.9 | 1e-130 | 40 | 412 | 1 | 348 |
XXXXXXXXXXXXXXXXXXXXXXX..XXXXX.XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX CS
EIN3 1 eelkkrmwkdqmllkrlkerkkqlledkeaatgakksnksneqarrkkmsraQDgiLkYMlkemevcnaqGfvYgiipekgkpvegasdsLr 92
eel++rmwkd+++lkr++er+k + +++++++k ++ +qarrkkmsraQDgiLkYMlk mevcn++GfvYgiipekgkpv+gasd++r
Aqcoe5G214000.1.p 40 EELERRMWKDRIKLKRIRERQKLVV---QRDAEKAKPKQNLDQARRKKMSRAQDGILKYMLKLMEVCNVRGFVYGIIPEKGKPVSGASDNIR 128
8****************99999765...3336899999****************************************************** PP
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX....XX----STTS-HHHHHHHHHHHSSSSSS-TTS--TTT--HHHH---S--HHHHHH CS
EIN3 93 aWWkekvefdrngpaaiskyqaknlilsgesslqtersseshslselqDTtlgSLLsalmqhcdppqrrfplekgvepPWWPtGkelwwgel 184
aWWkekv+fd+ngpaai+ky+a++ + ++++++ + + ++sl++lqD+tlgSLLs+lmqhcdppqr++pl+kg++pPWWP+G+e+ww ++
Aqcoe5G214000.1.p 129 AWWKEKVKFDKNGPAAIAKYDAECFAIGEAHRK--KDGNCKSSLQDLQDATLGSLLSSLMQHCDPPQRKYPLDKGIPPPWWPSGTEDWWIKM 218
***************************988888..6899999************************************************** PP
T--TT--.-----GGG--HHHHHHHHHHHHHHTGGGHHHHHHTTTTSSSSTTT--SHHHHHHHHHHTTTTT-S--XXXXXX........... CS
EIN3 185 glskdqgtppykkphdlkkawkvsvLtavikhmsptieeirelerqskylqdkmsakesfallsvlnqeekecatvsahss........... 265
gl+k q ppykkphdlkk+wkv+vLtavikhmsp+i++i++++rqsk+lqdkm+akes+++l vl+ ee+ ++++ s
Aqcoe5G214000.1.p 219 GLPKGQI-PPYKKPHDLKKMWKVGVLTAVIKHMSPDIAKIKRHVRQSKCLQDKMTAKESAIWLGVLSCEESSLQRLVLASDsggsgipktpp 309
******9.9**************************************************************999988775545777766554 PP
XXXX..XXXXXXXXXXXXXXXXXXXXXX.XXXXXXXXXX.......................XXXXXXXXXXXXXXXXXXXXX......XXX CS
EIN3 266 slrk..qspkvtlsceqkedvegkkeskikhvqavktta.......................gfpvvrkrkkkpsesakvsskevsrtcqss 332
++++vt+++++++dv+g ++ + ++++++ + +++ + +rk+++ +++ v++ ++
Aqcoe5G214000.1.p 310 G--GfgERKEVTANSDSDYDVDGADDGAGSVSSNNDKSNellaieprrdlpnnpnrsvqnkeQVEEQPRRKRQRPRETRVNQPV---VSNHT 396
2..145999*************88888866555555545799999999******999999999999999998888888877663...57778 PP
XXXX.XXXXXXXXXXX CS
EIN3 333 qfrgsetelifadkns 348
++rg+e++ ++d+n
Aqcoe5G214000.1.p 397 ELRGEEPRSALPDMNL 412
8999999999999886 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04873 | 1.5E-122 | 40 | 286 | No hit | No description |
| Gene3D | G3DSA:1.10.3180.10 | 1.3E-68 | 163 | 293 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| SuperFamily | SSF116768 | 1.57E-59 | 168 | 288 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042762 | Biological Process | regulation of sulfur metabolic process | ||||
| GO:0071281 | Biological Process | cellular response to iron ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 609 aa Download sequence Send to blast |
MENFMMDANE FGDNSDIEVD DLRDESMADK DVSDEEIDAE ELERRMWKDR IKLKRIRERQ 60 KLVVQRDAEK AKPKQNLDQA RRKKMSRAQD GILKYMLKLM EVCNVRGFVY GIIPEKGKPV 120 SGASDNIRAW WKEKVKFDKN GPAAIAKYDA ECFAIGEAHR KKDGNCKSSL QDLQDATLGS 180 LLSSLMQHCD PPQRKYPLDK GIPPPWWPSG TEDWWIKMGL PKGQIPPYKK PHDLKKMWKV 240 GVLTAVIKHM SPDIAKIKRH VRQSKCLQDK MTAKESAIWL GVLSCEESSL QRLVLASDSG 300 GSGIPKTPPG GFGERKEVTA NSDSDYDVDG ADDGAGSVSS NNDKSNELLA IEPRRDLPNN 360 PNRSVQNKEQ VEEQPRRKRQ RPRETRVNQP VVSNHTELRG EEPRSALPDM NLSDLQSAGK 420 QMDAIQYENA PVPLEKGLDG LPGLPESAPQ PVSVLPSSNV ATTQSMFVAG RPLLYPIMQV 480 PELHPPPPND FHNMSGTYGI PLGSQQQGMA IVDPQIRPEE NGVNVLGLHG TMNGATVGEM 540 HHYTKGTFES EQDKPLENHF GSPFDGFSFE FGDFGSPFNF DHGIDATSSF DTVDVDLLDD 600 DIIQFFGS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wij_A | 3e-71 | 170 | 292 | 12 | 134 | ETHYLENE-INSENSITIVE3-like 3 protein |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 376 | 382 | RKRQRPR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may be involved in the ethylene response pathway. {ECO:0000269|PubMed:9215635, ECO:0000269|PubMed:9851977}. | |||||
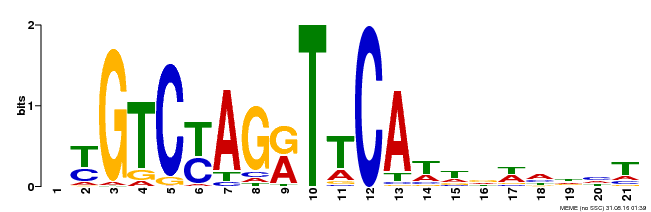
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00233 | DAP | Transfer from AT1G73730 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019053325.1 | 0.0 | PREDICTED: ETHYLENE INSENSITIVE 3-like 3 protein isoform X2 | ||||
| Swissprot | O23116 | 1e-172 | EIL3_ARATH; ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| TrEMBL | A0A2G5EI43 | 0.0 | A0A2G5EI43_AQUCA; Uncharacterized protein | ||||
| STRING | Aquca_007_00017.1 | 0.0 | (Aquilegia coerulea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73730.1 | 1e-155 | ETHYLENE-INSENSITIVE3-like 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aqcoe5G214000.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



