 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aqcoe5G381600.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Ranunculales; Ranunculaceae; Thalictroideae; Aquilegia
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 1545aa MW: 172277 Da PI: 9.118 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 11.1 | 0.0013 | 1429 | 1454 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
y+C C++ Fs+k +L+ H ++ +
Aqcoe5G381600.1.p 1429 YQCDldGCTMGFSSKQELVLHKKNiC 1454
99*******************99866 PP
| |||||||
| 2 | zf-C2H2 | 11.1 | 0.0013 | 1454 | 1476 | 3 | 23 |
ET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 3 Cp..dCgksFsrksnLkrHirtH 23
C+ Cgk F ++ +L++H r+H
Aqcoe5G381600.1.p 1454 CSvkGCGKKFFSHKYLVQHRRVH 1476
77779****************99 PP
| |||||||
| 3 | zf-C2H2 | 11.2 | 0.0011 | 1512 | 1538 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH..T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt..H 23
y+C Cg++F+ s++ rH r+ H
Aqcoe5G381600.1.p 1512 YVCRekGCGQTFRFVSDFSRHKRKtgH 1538
99********************99666 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00545 | 3.9E-16 | 35 | 76 | IPR003349 | JmjN domain |
| PROSITE profile | PS51183 | 14.659 | 36 | 77 | IPR003349 | JmjN domain |
| Pfam | PF02375 | 7.9E-15 | 37 | 70 | IPR003349 | JmjN domain |
| SMART | SM00558 | 1.6E-48 | 217 | 386 | IPR003347 | JmjC domain |
| PROSITE profile | PS51184 | 33.982 | 220 | 386 | IPR003347 | JmjC domain |
| SuperFamily | SSF51197 | 1.02E-26 | 231 | 382 | No hit | No description |
| Pfam | PF02373 | 2.8E-36 | 250 | 369 | IPR003347 | JmjC domain |
| SMART | SM00355 | 12 | 1429 | 1451 | IPR015880 | Zinc finger, C2H2-like |
| SMART | SM00355 | 0.015 | 1452 | 1476 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 12.487 | 1452 | 1481 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 3.0E-5 | 1453 | 1480 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 7.97E-6 | 1453 | 1488 | No hit | No description |
| PROSITE pattern | PS00028 | 0 | 1454 | 1476 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 2.2E-8 | 1481 | 1506 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0014 | 1482 | 1506 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 11.177 | 1482 | 1511 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1484 | 1506 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 1.33E-9 | 1492 | 1536 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 3.3E-9 | 1507 | 1535 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.38 | 1512 | 1538 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.034 | 1512 | 1543 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1514 | 1538 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009826 | Biological Process | unidimensional cell growth | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0033169 | Biological Process | histone H3-K9 demethylation | ||||
| GO:0035067 | Biological Process | negative regulation of histone acetylation | ||||
| GO:0040010 | Biological Process | positive regulation of growth rate | ||||
| GO:0045815 | Biological Process | positive regulation of gene expression, epigenetic | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0071558 | Molecular Function | histone demethylase activity (H3-K27 specific) | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1545 aa Download sequence Send to blast |
MAGNHSTIGG GGGGGGIVVA EVVHEVPQWL KNLPIAPEFH PTLAEFEDPI SYILKIEKQA 60 SQYGICKIVP PVSLSSKKTT ISNLNRSFSS PSSRYNNNNN NHIHNPNSSI SSPTFTTRFQ 120 QIGFDPRKGR PVLKPVWQSG EYYTLQQFET KAKQFEKIHL KKCGKKGGYS SLEIETLFWR 180 ASVDKPFTVE YANDMPGSAF GPVKLGKTFK ETGEGMTVGD TAWNMRGVAR SKGSLLRFMK 240 EEIPGVTSPM VYIAMLFSWF AWHVEDHDLH SLNYMHMGAS KTWYGVPKDA AFAFQEVVRA 300 HGYGGDFNPL VTFATLGEKT TVMSPEVLIG AGIPCCRLVQ NVGEFVVTFP GAYHSGFSHG 360 FNCAEASNIA TPEWLRYAKD AAIRRASVNC PPMVSHFQLL YALAMTLCSR MPMTIGDEPR 420 SSRLKDKKKG EGEGMVKEFF VQSVMQNNDL LHVLSSHGSS CVLMPRNSSE ISIYLNRRIG 480 IQKKVKRRMS LGLCSNSEAS KAARVLHPDE DTLERKMRLN QLTAFNAVND TSNSLFMEDK 540 LSSLAVKHNH QDLCNSAIVG NDMKNNCTSQ DAVLLEQGRF PCVQCGILSY ACVAVIEPKE 600 AAAQYLMSAD CSFFNDWKAG SGVLKEESTA VTENEKILER NYTLGVGVID NDYKDDLYDF 660 SVQSGDCQVE DQEVDVASHT GPQKGISSLA LLASAYGNSS DSDEELAAPK MVVNDDDDDI 720 RTRKLLGDTN VFSPSSDLCD GFTLHTVDPS QHDGASGSIP HSSSNQGRGN EVTLRMARCT 780 SSSRRKRRWT SSEVKDRGEL TSDYPLESDV ETLVPVNMKR LKDSYRDEED VSGGVIAQST 840 EIHETDLYTN QVETDPADSI VPLSSFCSGS AFSQVPLSET TNISIQSTSM SLMHISREES 900 SRKHIFCLEH AVEVEKQLRP IGGAHILLIC HPEYPRVEAQ AKSLTQDLGI DHLWKDISYR 960 EATKEDQERI GLALDDEEAD PSNGDWSVKL GINLYYSVHL SKSPLYTKQM PYNSIIYKAF 1020 GCSSMNNPME VKVSGKVPRQ KKIVVAGKWC GKVWMSNQVH PYLVQREMVE EDPMNLLVQT 1080 TPFLRPRSKL ERKVEINQLL KEPLISGKRS APEKTTIARK LGQKKKRKFG KAATKKQKSA 1140 ELESPTQTEE ESCESDSSEK KSLRVTTTVA RKLGQMKKRN FGKVAAKKQK SPELESPTHT 1200 PEESSESDSS EKKSSPRVRA TVARKLGQLE KRKVGKVAAK KQKSPELEST QTAEDSSESD 1260 SYPSSNGRFL STWGTKRPVQ QSSVKNIRQY DSNKKVEIKG EPTMAIKLEQ KKRMFGKEAA 1320 KKPKSPQLES PTQSEEDSSE SDSSPLNNEK FLKTGRTKNE RPLLQYSRQY DSKPKIEIEG 1380 GPSTRLRRRP SKPVEESKVK PVGRKQINTK KGKKALPNSN DIKSEEADYQ CDLDGCTMGF 1440 SSKQELVLHK KNICSVKGCG KKFFSHKYLV QHRRVHLDDR PLKCPWKGCK MTFKWAWART 1500 EHIRVHTGDR PYVCREKGCG QTFRFVSDFS RHKRKTGHSV KGKG* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6ip0_A | 2e-71 | 28 | 407 | 7 | 353 | Transcription factor jumonji (Jmj) family protein |
| 6ip4_A | 2e-71 | 28 | 407 | 7 | 353 | Arabidopsis JMJ13 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00608 | ChIP-seq | Transfer from AT3G48430 | Download |
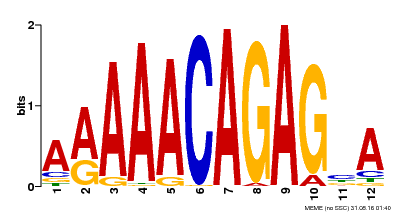
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| TrEMBL | A0A2G5E6S5 | 0.0 | A0A2G5E6S5_AQUCA; Uncharacterized protein (Fragment) | ||||
| STRING | Aquca_011_00361.1 | 0.0 | (Aquilegia coerulea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48430.1 | 0.0 | relative of early flowering 6 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aqcoe5G381600.1.p |



