 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aqcoe6G001700.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Ranunculales; Ranunculaceae; Thalictroideae; Aquilegia
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 374aa MW: 42540.2 Da PI: 7.3403 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 159.3 | 1.6e-49 | 23 | 148 | 2 | 128 |
NAM 2 ppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpkkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevl 93
+pGfrFhPt+eel+ +yL++kvegk++++ e i+ +d+y+++Pw+Lp+ ++ +ekew+f+++rd+ky++g+r+nr+t+sgyWkatg d+ +
Aqcoe6G001700.1.p 23 MPGFRFHPTEEELIEFYLRRKVEGKRFNV-ELITFLDLYRYDPWELPALAAIGEKEWFFYVPRDRKYRNGDRPNRVTTSGYWKATGADRMIR 113
79***************************.89***************8778899************************************** PP
NAM 94 skkgelvglkktLvfykgrapkgektdWvmheyrl 128
+ + + +glkktLvfy+g+apkg +t+W+m+eyrl
Aqcoe6G001700.1.p 114 NDNLRSIGLKKTLVFYSGKAPKGIRTSWIMNEYRL 148
*********************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51005 | 55.264 | 22 | 169 | IPR003441 | NAC domain |
| SuperFamily | SSF101941 | 3.79E-55 | 22 | 169 | IPR003441 | NAC domain |
| Pfam | PF02365 | 1.1E-24 | 24 | 148 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 374 aa Download sequence Send to blast |
MAIAATMSQN NDDKDNHDHD MVMPGFRFHP TEEELIEFYL RRKVEGKRFN VELITFLDLY 60 RYDPWELPAL AAIGEKEWFF YVPRDRKYRN GDRPNRVTTS GYWKATGADR MIRNDNLRSI 120 GLKKTLVFYS GKAPKGIRTS WIMNEYRLPQ QDTDRYQKAE ISLCRVYKRP GVEDHARVPG 180 SSHSSAPSSS RGTQPDKRQQ NLITYGISSF QAYDSNNSPA TTTTTTTEME KLNEADASST 240 SNDTGTALGI ANVSPMIRSA VLSQASTIEE VSGTAFIQQY SRQMSSLPSN TCNTNTLFSY 300 PPIIPTADDL RRLVDYQHAY NQQHHQQQYI NHQHCTQFMP SSSSNSLPNS SMPTFSADKL 360 WEWNPIPDSN RDN* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 3e-54 | 24 | 169 | 19 | 165 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 3e-54 | 24 | 169 | 19 | 165 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 3e-54 | 24 | 169 | 19 | 165 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 3e-54 | 24 | 169 | 19 | 165 | NO APICAL MERISTEM PROTEIN |
| 3swm_A | 4e-54 | 24 | 169 | 22 | 168 | NAC domain-containing protein 19 |
| 3swm_B | 4e-54 | 24 | 169 | 22 | 168 | NAC domain-containing protein 19 |
| 3swm_C | 4e-54 | 24 | 169 | 22 | 168 | NAC domain-containing protein 19 |
| 3swm_D | 4e-54 | 24 | 169 | 22 | 168 | NAC domain-containing protein 19 |
| 3swp_A | 4e-54 | 24 | 169 | 22 | 168 | NAC domain-containing protein 19 |
| 3swp_B | 4e-54 | 24 | 169 | 22 | 168 | NAC domain-containing protein 19 |
| 3swp_C | 4e-54 | 24 | 169 | 22 | 168 | NAC domain-containing protein 19 |
| 3swp_D | 4e-54 | 24 | 169 | 22 | 168 | NAC domain-containing protein 19 |
| 4dul_A | 3e-54 | 24 | 169 | 19 | 165 | NAC domain-containing protein 19 |
| 4dul_B | 3e-54 | 24 | 169 | 19 | 165 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that acts as a floral repressor. Controls flowering time by negatively regulating CONSTANS (CO) expression in a GIGANTEA (GI)-independent manner. Regulates the plant cold response by positive regulation of the cold response genes COR15A and KIN1. May coordinate cold response and flowering time. {ECO:0000269|PubMed:17653269}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00257 | DAP | Transfer from AT2G02450 | Download |
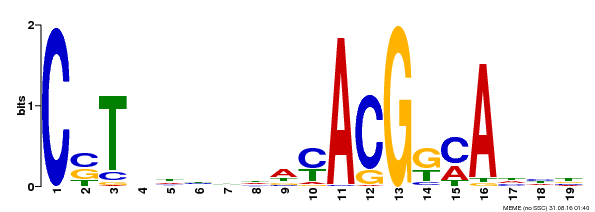
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Circadian regulation with a peak of expression at dawn under continuous light conditions (PubMed:17653269). Circadian regulation with a peak of expression around dusk and lowest expression around dawn under continuous light conditions (at protein level) (PubMed:17653269). {ECO:0000269|PubMed:17653269}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010252261.1 | 1e-153 | PREDICTED: NAC domain-containing protein 35 | ||||
| Swissprot | Q9ZVP8 | 1e-109 | NAC35_ARATH; NAC domain-containing protein 35 | ||||
| TrEMBL | A0A2G5DU09 | 0.0 | A0A2G5DU09_AQUCA; Uncharacterized protein | ||||
| STRING | Aquca_014_00018.1 | 0.0 | (Aquilegia coerulea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G02450.1 | 1e-108 | NAC domain containing protein 35 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aqcoe6G001700.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



