 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aqcoe6G003100.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Ranunculales; Ranunculaceae; Thalictroideae; Aquilegia
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 513aa MW: 56976.7 Da PI: 6.681 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 39.7 | 9e-13 | 365 | 411 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCC.C...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPk.askapskKlsKaeiLekAveYIksLq 55
+h e+Er+RR+++N++f Lr ++P+ + K++Ka+ L A+ YI +Lq
Aqcoe6G003100.1.p 365 NHVEAERQRREKLNQRFYALRAVVPNiS------KMDKASLLGDAITYITDLQ 411
799***********************66......*****************99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 4.3E-43 | 48 | 225 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| PROSITE profile | PS50888 | 16.831 | 361 | 410 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 3.11E-14 | 364 | 415 | No hit | No description |
| SuperFamily | SSF47459 | 1.83E-18 | 364 | 430 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 3.0E-10 | 365 | 411 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 4.7E-17 | 365 | 425 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 3.1E-15 | 367 | 416 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 513 aa Download sequence Send to blast |
MGEKGWVSDE DKAMFESVLS SEALRFLINS SSNGGAIPKE SMVSQSVKQE LRRLVDGFNW 60 SYAILWQVSR SNSGEPNLIW GGGHCNRSNV EQQQQQEVKG FTHSQELKKK LLQYLHCLFG 120 GSANSGERLD SVSDVEFFYL TSMYYSLPFD DAPTGLAWSF ASNRTIWASD QNNCMGHYNS 180 RAFLAKSTGI RTLVCVPIQS GVVELGSINQ FPEDQSVLQM IKNVFSGSRS ITASTSASAP 240 KIFGHDLSLA KPKPRSLTIN FSPKLEEEED TEFSLVSYTN QSLVPGHSNA LVDQAFGTFS 300 NGSRIDDNSE EKLFLQGNHS CPGNLNPQPR MLSSDQSSMD VSRSDERKPR KRGRKPANGR 360 EEPLNHVEAE RQRREKLNQR FYALRAVVPN ISKMDKASLL GDAITYITDL QTKIRVLETE 420 KEIARNSSPQ QSPIPEIDVQ TRQEDTVVQV SCPLDAHPVS RVLRAFQETQ VTVNQSTVST 480 IDNDSVLHTF SIRTQGGASE ILKERLVAAL SR* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5gnj_A | 1e-26 | 359 | 426 | 2 | 69 | Transcription factor MYC2 |
| 5gnj_B | 1e-26 | 359 | 426 | 2 | 69 | Transcription factor MYC2 |
| 5gnj_E | 1e-26 | 359 | 426 | 2 | 69 | Transcription factor MYC2 |
| 5gnj_F | 1e-26 | 359 | 426 | 2 | 69 | Transcription factor MYC2 |
| 5gnj_G | 1e-26 | 359 | 426 | 2 | 69 | Transcription factor MYC2 |
| 5gnj_I | 1e-26 | 359 | 426 | 2 | 69 | Transcription factor MYC2 |
| 5gnj_M | 1e-26 | 359 | 426 | 2 | 69 | Transcription factor MYC2 |
| 5gnj_N | 1e-26 | 359 | 426 | 2 | 69 | Transcription factor MYC2 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 346 | 354 | RKPRKRGRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that negatively regulates jasmonate (JA) signaling (PubMed:30610166). Negatively regulates JA-dependent response to wounding, JA-induced expression of defense genes, JA-dependent responses against herbivorous insects, and JA-dependent resistance against Botrytis cinerea infection (PubMed:30610166). Plays a positive role in resistance against the bacterial pathogen Pseudomonas syringae pv tomato DC3000 (PubMed:30610166). {ECO:0000269|PubMed:30610166}. | |||||
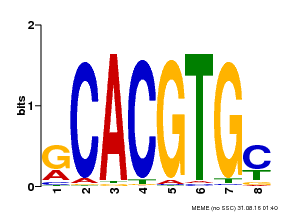
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00099 | PBM | Transfer from AT4G16430 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by wounding, feeding with herbivorous insects, infection with the fungal pathogen Botrytis cinerea and infection with the bacterial pathogen Pseudomonas syringae pv tomato DC3000. {ECO:0000269|PubMed:30610166}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010252341.1 | 0.0 | PREDICTED: transcription factor bHLH3 | ||||
| Refseq | XP_010252342.1 | 0.0 | PREDICTED: transcription factor bHLH3 | ||||
| Swissprot | A0A3Q7H216 | 1e-151 | MTB3_SOLLC; Transcription factor MTB3 | ||||
| TrEMBL | A0A2G5DU52 | 0.0 | A0A2G5DU52_AQUCA; Uncharacterized protein | ||||
| STRING | Aquca_014_00031.1 | 0.0 | (Aquilegia coerulea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16430.1 | 1e-136 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aqcoe6G003100.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



