 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aqcoe6G063200.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Ranunculales; Ranunculaceae; Thalictroideae; Aquilegia
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 385aa MW: 42489.4 Da PI: 6.9418 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 64.8 | 1.7e-20 | 172 | 221 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
y+GVr+++ +g+WvAeIr p++ r+r +lg+f+taeeAa a+++a+ kl+g
Aqcoe6G063200.1.p 172 LYRGVRQRH-WGKWVAEIRLPRN---RTRLWLGTFDTAEEAALAYDKAAYKLRG 221
59******9.**********955...5*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 5.92E-34 | 171 | 231 | No hit | No description |
| PROSITE profile | PS51032 | 23.248 | 172 | 229 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 2.8E-38 | 172 | 235 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 3.1E-32 | 172 | 230 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 5.04E-23 | 172 | 230 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 2.7E-14 | 173 | 221 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.9E-11 | 173 | 184 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.9E-11 | 195 | 211 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009736 | Biological Process | cytokinin-activated signaling pathway | ||||
| GO:0009873 | Biological Process | ethylene-activated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0045595 | Biological Process | regulation of cell differentiation | ||||
| GO:0071472 | Biological Process | cellular response to salt stress | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 385 aa Download sequence Send to blast |
MAAALDFYSR SNSNGSTLTD PCSEELMMAL QPFITSASPT QNPSPSSSST ISFDSSSPSP 60 YPFSSFESNL CNPVYCSTST TPMFSQGLSS YDQVGLEQTG SIGLNNLTSS QIHQIQTQIQ 120 FQQQQHMAAM AAAASIHNQQ LNQLQQQTTL SFLSPKAIPM KQVGSSSKST KLYRGVRQRH 180 WGKWVAEIRL PRNRTRLWLG TFDTAEEAAL AYDKAAYKLR GDLARLNFPH LRHQGSHVGS 240 EFGDYKPLHS SVDAKLEAIC ENMKQGNLGK PTVVLKKNAK KKTMSQKSSK TQTNVINKSV 300 ENLSNGEMSI LGSEDIKMES SLTTSQSEID VASSPESDIS RFQECTETPW NDSDNFALQK 360 YPSIDNYPLQ KYPSFDIDWD SILS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gcc_A | 4e-22 | 171 | 229 | 1 | 60 | ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 |
| 2gcc_A | 5e-22 | 171 | 229 | 4 | 63 | ATERF1 |
| 3gcc_A | 5e-22 | 171 | 229 | 4 | 63 | ATERF1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00026 | PBM | Transfer from AT1G78080 | Download |
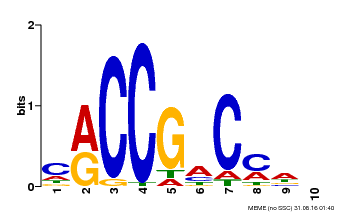
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010261262.1 | 1e-112 | PREDICTED: ethylene-responsive transcription factor RAP2-4-like | ||||
| TrEMBL | A0A2G5DX49 | 0.0 | A0A2G5DX49_AQUCA; Uncharacterized protein | ||||
| STRING | Aquca_014_00592.1 | 0.0 | (Aquilegia coerulea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G78080.1 | 4e-57 | related to AP2 4 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aqcoe6G063200.1.p |



