 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aqcoe6G257900.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Ranunculales; Ranunculaceae; Thalictroideae; Aquilegia
|
||||||||
| Family | SRS | ||||||||
| Protein Properties | Length: 322aa MW: 34509.9 Da PI: 8.3377 | ||||||||
| Description | SRS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF702 | 226.1 | 6.5e-70 | 99 | 255 | 3 | 154 |
DUF702 3 sgtasCqdCGnqakkdCaheRCRtCCksrgfdCathvkstWvpaakrrerqqqlaaasskaaasa......aeaaskrkrelkskkqsalss 88
+g++sCqdCGnqakkdC+h+RCRtCCksrgf+C+thvkstWvpaa+rrerqqql+++++++++++ +++++kr re+ ++s+l++
Aqcoe6G257900.1.p 99 VGGKSCQDCGNQAKKDCVHMRCRTCCKSRGFQCSTHVKSTWVPAARRRERQQQLQSLQQQQQQQHqllqhhHHQNPKRLREN--PSSSSLAC 188
6789****************************************************99999888778854333444444443..33333444 PP
DUF702 89 tkls.saeskkeletsslPeevsseavfrcvrvssvddgeeelaYqtavsigGhvfkGiLydqGlee 154
t+++ ++++ le+ ++P+ev+s+avfrcvrvs+++d+e+e+aYqtav+igGhvfkG+LydqG e+
Aqcoe6G257900.1.p 189 TTTTrLPSTTTGLEVGNFPSEVNSQAVFRCVRVSGMNDAEDEYAYQTAVNIGGHVFKGLLYDQGSED 255
33330344566789999***********************************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05142 | 7.6E-68 | 101 | 254 | IPR007818 | Protein of unknown function DUF702 |
| TIGRFAMs | TIGR01623 | 8.9E-28 | 103 | 145 | IPR006510 | Zinc finger, lateral root primordium type 1 |
| TIGRFAMs | TIGR01624 | 2.5E-26 | 206 | 252 | IPR006511 | Lateral Root Primordium type 1, C-terminal |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010051 | Biological Process | xylem and phloem pattern formation | ||||
| GO:0010252 | Biological Process | auxin homeostasis | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0048479 | Biological Process | style development | ||||
| GO:0048480 | Biological Process | stigma development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0046982 | Molecular Function | protein heterodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 322 aa Download sequence Send to blast |
MAGFSLGSGG GGGGGNPSSD IPPESFFLYR NEEIAAFNTS KGFELWQQHL QRQQRYHQEI 60 YSSADVSDES SSRGAAANIG HVSMMMRSQG ASGSGGGSVG GKSCQDCGNQ AKKDCVHMRC 120 RTCCKSRGFQ CSTHVKSTWV PAARRRERQQ QLQSLQQQQQ QQHQLLQHHH HQNPKRLREN 180 PSSSSLACTT TTRLPSTTTG LEVGNFPSEV NSQAVFRCVR VSGMNDAEDE YAYQTAVNIG 240 GHVFKGLLYD QGSEDRYVPG ESSSGGGGRH LIAGTTAATN TTAANTSNAA ASLLDPSSSQ 300 LYPAPLTAFM AGTQFFPHPR S* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator involved in the transcriptional regulation of terpene biosynthesis in glandular trichomes (PubMed:24884371, PubMed:24142382). Binds to the promoter of the linalool synthase TPS5 and promotes TPS5 gene transactivation (PubMed:24884371, PubMed:24142382). Acts synergistically with MYC1 in the transactivation of TPS5 (PubMed:24884371). {ECO:0000269|PubMed:24142382, ECO:0000269|PubMed:24884371}. | |||||
| UniProt | Transcription activator that binds DNA on 5'-ACTCTAC-3' and promotes auxin homeostasis-regulating gene expression (e.g. YUC genes), as well as genes affecting stamen development, cell expansion and timing of flowering. Synergistically with other SHI-related proteins, regulates gynoecium, stamen and leaf development in a dose-dependent manner, controlling apical-basal patterning. Promotes style and stigma formation, and influences vascular development during gynoecium development. May also have a role in the formation and/or maintenance of the shoot apical meristem (SAM). {ECO:0000269|PubMed:12361963, ECO:0000269|PubMed:16740145, ECO:0000269|PubMed:16740146, ECO:0000269|PubMed:18811619, ECO:0000269|PubMed:20154152, ECO:0000269|PubMed:22318676}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00613 | PBM | Transfer from AT3G51060 | Download |
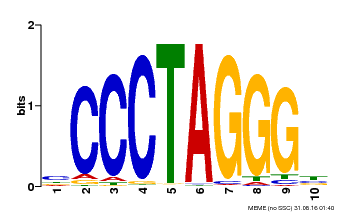
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Regulated by ESR1 and ESR2. {ECO:0000269|PubMed:21976484}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | CR955005 | 3e-43 | CR955005.2 Medicago truncatula chromosome 5 clone mte1-58c24, COMPLETE SEQUENCE. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010271515.1 | 1e-115 | PREDICTED: protein SHI RELATED SEQUENCE 1-like | ||||
| Swissprot | K4B6C9 | 4e-76 | EOT1_SOLLC; Protein EXPRESSION OF TERPENOIDS 1 | ||||
| Swissprot | Q9SD40 | 5e-76 | SRS1_ARATH; Protein SHI RELATED SEQUENCE 1 | ||||
| TrEMBL | A0A2G5EMS9 | 0.0 | A0A2G5EMS9_AQUCA; Uncharacterized protein | ||||
| STRING | Aquca_006_00068.1 | 0.0 | (Aquilegia coerulea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G51060.1 | 2e-61 | Lateral root primordium (LRP) protein-related | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aqcoe6G257900.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



