 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aqcoe6G325700.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Ranunculales; Ranunculaceae; Thalictroideae; Aquilegia
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 373aa MW: 42295.8 Da PI: 4.4629 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 34.2 | 5.7e-11 | 207 | 251 | 3 | 51 |
XXCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 3 elkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkelee 51
k+ +r ++NR AA rsR+RKk++++ Le k+k Leae +L+ +
Aqcoe6G325700.1.p 207 NTKKRKRQMRNRDAAVRSRERKKMYVKDLEVKSKYLEAECIRLQ----R 251
6799***********************************86554....3 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.20.5.170 | 2.0E-12 | 200 | 268 | No hit | No description |
| SMART | SM00338 | 1.7E-7 | 202 | 269 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 4.4E-9 | 207 | 251 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 9.887 | 207 | 251 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 1.63E-10 | 209 | 267 | No hit | No description |
| CDD | cd14704 | 7.46E-16 | 210 | 261 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 212 | 227 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0030968 | Biological Process | endoplasmic reticulum unfolded protein response | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005789 | Cellular Component | endoplasmic reticulum membrane | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 373 aa Download sequence Send to blast |
MIGDYGRGDV IEESDILNHD FDFDFDFSLF NDDEDLFADL FPYSSSSSCY DNHNESSFSS 60 LFRDIEDKLL NNDEKEYQRD ENTTTTDVDE FFSDLQLPSY FVHQHHPHSH SQSQDDQEIS 120 MDDLLVDDDV PPPSPGLNTN YSPSNSSSNQ VVTILETPDP QTSTPESQVS FQDDDDDHQE 180 KASLEKETTI TTTDSASLED KEDDDPNTKK RKRQMRNRDA AVRSRERKKM YVKDLEVKSK 240 YLEAECIRLQ RLLQCCAAEN HALHFQLQEK AFSASRTKQE SAVLFLESLL LGSLLWFLGI 300 LGLFSPEMLR VQLPADLLVN VENQKVVDLV ALGRAKSKSI GLGTLRLFFK SKRCKASRSK 360 MKNMICNGAV AA* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 209 | 228 | KRKRQMRNRDAAVRSRERKK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00019 | PBM | Transfer from AT1G42990 | Download |
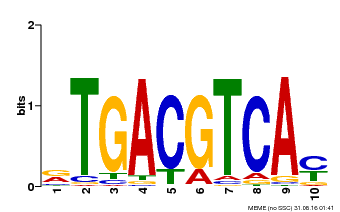
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| TrEMBL | A0A2G5C4S5 | 0.0 | A0A2G5C4S5_AQUCA; Uncharacterized protein | ||||
| STRING | Aquca_095_00028.1 | 0.0 | (Aquilegia coerulea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G42990.1 | 8e-16 | basic region/leucine zipper motif 60 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aqcoe6G325700.1.p |



