 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aqcoe7G053700.2.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Ranunculales; Ranunculaceae; Thalictroideae; Aquilegia
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 255aa MW: 28849.3 Da PI: 9.7567 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 98.8 | 2.2e-31 | 9 | 59 | 1 | 51 |
S---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE- CS
SRF-TF 1 krienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklyeyss 51
krienk+nrqvtfskRrng+lKKA+ELSvLCdaev +iifss+gklye++s
Aqcoe7G053700.2.p 9 KRIENKINRQVTFSKRRNGLLKKAYELSVLCDAEVGLIIFSSRGKLYEFAS 59
79***********************************************86 PP
| |||||||
| 2 | K-box | 104.8 | 1.1e-34 | 78 | 173 | 5 | 99 |
K-box 5 sgks.leeakaeslqqelakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkekelqeenkaLr 95
+g+ +++++++ ++qe++kLk+++e+Lqr+qRhllGedL++Ls+keL++Le+qLe +l + R++K+++++eq+e+l++ke+ l ++nk+L+
Aqcoe7G053700.2.p 78 DGTLaVADRETQGWYQEVSKLKAKYESLQRSQRHLLGEDLGPLSVKELHNLEKQLEGALAQARQRKTQVMMEQMEDLRRKERHLGDINKQLK 169
444468999*********************************************************************************** PP
K-box 96 kkle 99
+k++
Aqcoe7G053700.2.p 170 NKYQ 173
9975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00432 | 1.9E-41 | 1 | 60 | IPR002100 | Transcription factor, MADS-box |
| PROSITE profile | PS50066 | 33.613 | 1 | 61 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 1.39E-45 | 2 | 78 | No hit | No description |
| SuperFamily | SSF55455 | 2.75E-33 | 2 | 77 | IPR002100 | Transcription factor, MADS-box |
| PROSITE pattern | PS00350 | 0 | 3 | 57 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 7.6E-33 | 3 | 23 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 8.3E-27 | 10 | 57 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 7.6E-33 | 23 | 38 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 7.6E-33 | 38 | 59 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF01486 | 1.6E-31 | 86 | 172 | IPR002487 | Transcription factor, K-box |
| PROSITE profile | PS51297 | 17.01 | 88 | 180 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009553 | Biological Process | embryo sac development | ||||
| GO:0009911 | Biological Process | positive regulation of flower development | ||||
| GO:0010094 | Biological Process | specification of carpel identity | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0010582 | Biological Process | floral meristem determinacy | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0048455 | Biological Process | stamen formation | ||||
| GO:0048459 | Biological Process | floral whorl structural organization | ||||
| GO:0048509 | Biological Process | regulation of meristem development | ||||
| GO:0048833 | Biological Process | specification of floral organ number | ||||
| GO:0080060 | Biological Process | integument development | ||||
| GO:0080112 | Biological Process | seed growth | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 255 aa Download sequence Send to blast |
MGRGRVELKR IENKINRQVT FSKRRNGLLK KAYELSVLCD AEVGLIIFSS RGKLYEFASA 60 GMSRTLERYQ RSSYNSQDGT LAVADRETQG WYQEVSKLKA KYESLQRSQR HLLGEDLGPL 120 SVKELHNLEK QLEGALAQAR QRKTQVMMEQ MEDLRRKERH LGDINKQLKN KYQLDAEGQA 180 PYRALQGSWE SNALVASNNF SMHASQSSSM DCEPTLQIGY HQFVSPEGGT SIPRTSAGEN 240 NNNNNNNFTQ GWAL* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5f28_A | 1e-22 | 1 | 75 | 1 | 73 | MEF2C |
| 5f28_B | 1e-22 | 1 | 75 | 1 | 73 | MEF2C |
| 5f28_C | 1e-22 | 1 | 75 | 1 | 73 | MEF2C |
| 5f28_D | 1e-22 | 1 | 75 | 1 | 73 | MEF2C |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in flower development. {ECO:0000250|UniProtKB:Q0HA25}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00315 | DAP | Transfer from AT2G45650 | Download |
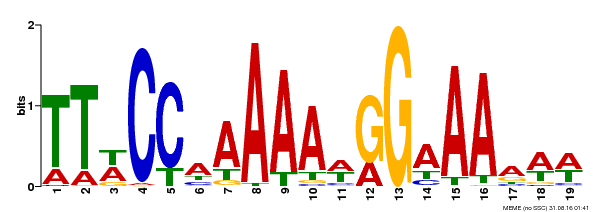
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | JX680248 | 0.0 | JX680248.1 Aquilegia coerulea MADS-box protein AGL6 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_026409535.1 | 1e-140 | MADS-box transcription factor 6-like | ||||
| Swissprot | Q8LLR1 | 1e-125 | MADS3_VITVI; Agamous-like MADS-box protein MADS3 | ||||
| TrEMBL | A0A2G5E1Z5 | 0.0 | A0A2G5E1Z5_AQUCA; Uncharacterized protein | ||||
| STRING | Aquca_013_00477.1 | 1e-179 | (Aquilegia coerulea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G45650.1 | 3e-96 | AGAMOUS-like 6 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aqcoe7G053700.2.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



