 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aqcoe7G157600.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Ranunculales; Ranunculaceae; Thalictroideae; Aquilegia
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 842aa MW: 92073.6 Da PI: 6.3298 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 58.4 | 1.2e-18 | 18 | 76 | 3 | 57 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHC....TS-HHHHHHHHHHHHHHHHC CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkkl....gLterqVkvWFqNrRakekk 57
k ++t+eq+e+Le+l++++++ps +r++L +++ +++ +q+kvWFqNrR +ek+
Aqcoe7G157600.1.p 18 KYVRYTPEQVEALERLYHECPKPSSIRRQQLIRECpilsNIEPKQIKVWFQNRRCREKQ 76
5679*****************************************************97 PP
| |||||||
| 2 | START | 169.2 | 2.8e-53 | 164 | 372 | 2 | 205 |
HHHHHHHHHHHHHHHC-TT-EEEEEXCCTTEEEEEEESSS.SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-SEEEEEEEECTT..EEE CS
START 2 laeeaaqelvkkalaeepgWvkssesengdevlqkfeeskvdsgealrasgvvdmvlallveellddkeqWdetlakaetlevissg..gal 91
+aee+++e+++ka+ ++ Wv+++ +++g++++ +++ s++++g a+ra+g+v +++ v+e+l+d++ W ++++ +++++v+ +g g++
Aqcoe7G157600.1.p 164 IAEETLTEFLSKATGTAVEWVQMPGMKPGPDSIGIIAISHGCPGVAARACGLVGLEPT-RVAEILKDRPSWFRDCRVVDVMNVLPTGtgGTI 254
789*******************************************************.8888888888*********************** PP
EEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--....-TTSEE-EESSEEEEEEEECTCEEEEEEEE-EE--SSXXHH CS
START 92 qlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppe...sssvvRaellpSgiliepksnghskvtwvehvdlkgrlphw 179
+l +++l+a+++l++ Rdf+ +Ry+ l++g++v++++S++s+q p+ +++vRae+lpSg+l++p+++g+s +++v+h+dl+ +++++
Aqcoe7G157600.1.p 255 ELLYMQLYAPTTLASaRDFWLLRYTSVLEDGSLVVCERSLSSTQGGPSmplVQHFVRAEMLPSGYLVRPCEGGGSIIHIVDHMDLEPSSVPE 346
***********************************************999999*************************************** PP
HHHHHHHHHHHHHHHHHHHHTXXXXX CS
START 180 llrslvksglaegaktwvatlqrqce 205
+lr+l++s ++ ++kt++a+l+++++
Aqcoe7G157600.1.p 347 VLRPLYESPTVLAQKTTMAALRQLRQ 372
**********************9876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 1.0E-18 | 7 | 76 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 15.742 | 13 | 77 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.2E-15 | 15 | 81 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 5.13E-17 | 17 | 81 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 9.37E-17 | 18 | 78 | No hit | No description |
| Pfam | PF00046 | 3.0E-16 | 19 | 76 | IPR001356 | Homeobox domain |
| CDD | cd14686 | 1.53E-6 | 70 | 109 | No hit | No description |
| PROSITE profile | PS50848 | 24.064 | 154 | 354 | IPR002913 | START domain |
| CDD | cd08875 | 4.59E-77 | 158 | 374 | No hit | No description |
| SuperFamily | SSF55961 | 1.22E-35 | 163 | 373 | No hit | No description |
| Gene3D | G3DSA:3.30.530.20 | 1.3E-20 | 163 | 369 | IPR023393 | START-like domain |
| SMART | SM00234 | 3.7E-38 | 163 | 373 | IPR002913 | START domain |
| Pfam | PF01852 | 7.4E-51 | 164 | 372 | IPR002913 | START domain |
| Pfam | PF08670 | 1.0E-52 | 698 | 840 | IPR013978 | MEKHLA |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0010014 | Biological Process | meristem initiation | ||||
| GO:0010075 | Biological Process | regulation of meristem growth | ||||
| GO:0010087 | Biological Process | phloem or xylem histogenesis | ||||
| GO:0048263 | Biological Process | determination of dorsal identity | ||||
| GO:0080060 | Biological Process | integument development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 842 aa Download sequence Send to blast |
MMAITSCKDG KGQMDNGKYV RYTPEQVEAL ERLYHECPKP SSIRRQQLIR ECPILSNIEP 60 KQIKVWFQNR RCREKQRKEA SRLQAVNRKL SAMNKLLMEE NDRLQKQVSQ LVYENGYFRQ 120 QPQTTAAALA TTDTSCESVV TSGQHHLTPQ HPPRDASPAG LLSIAEETLT EFLSKATGTA 180 VEWVQMPGMK PGPDSIGIIA ISHGCPGVAA RACGLVGLEP TRVAEILKDR PSWFRDCRVV 240 DVMNVLPTGT GGTIELLYMQ LYAPTTLASA RDFWLLRYTS VLEDGSLVVC ERSLSSTQGG 300 PSMPLVQHFV RAEMLPSGYL VRPCEGGGSI IHIVDHMDLE PSSVPEVLRP LYESPTVLAQ 360 KTTMAALRQL RQIALEASQS TVTGWGRRPA ALRALSQRLS RGFNEALNGF TDDGWSPMGS 420 DGMDDVTILV NSSPGKMSGV NIAYGNGFPA VTSGILCAKA SMLLQNVPPP ILLRFLREHR 480 SEWADSNIDA YSAAALKSGP CILPGSRVGG FGGQVILPLA HTIEHEEFLE VIKLESIGHC 540 QEDTLMSRDM FLLQLCSGVD ENAVGTCAEL VFAPIDSSFA DEAPLLPSGF RIIPLDFGVD 600 ASSPNRTLDL ASALEIGPAG NKVPGDYSGS CGSMRSVMTI AFQFAFESHL QENVASMARQ 660 YLRSIISSVQ RVALALSPSL LSPHSGLRPP PGTPEARTLA RWICQSYRCY LGVELLKPTT 720 DGSESNLKTL WHHSDAIMCC SLKALPVFTF ANQAGLDMLE TTLVALQDIT LEKIFDDHGR 780 KTLCSEFPQI MQQGFACLQG GICLSSMGRP VSYERAVAWK VLNEEENAHC ICFMFVNWSF 840 V* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of meristem development to promote lateral organ formation. May regulates procambial and vascular tissue formation or maintenance, and vascular development in inflorescence stems. {ECO:0000269|PubMed:15598805, ECO:0000269|PubMed:15705957, ECO:0000269|PubMed:16617092}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00200 | DAP | Transfer from AT1G52150 | Download |
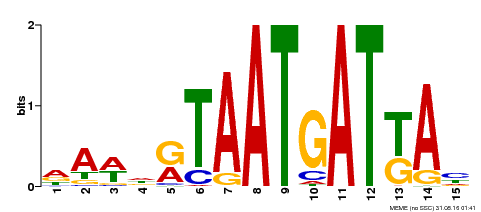
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By auxin. Repressed by miR165 and miR166. {ECO:0000269|PubMed:15773855, ECO:0000269|PubMed:16033795, ECO:0000269|PubMed:16617092, ECO:0000269|PubMed:17237362}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010267183.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein ATHB-15-like | ||||
| Refseq | XP_010267184.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein ATHB-15-like | ||||
| Swissprot | Q9ZU11 | 0.0 | ATB15_ARATH; Homeobox-leucine zipper protein ATHB-15 | ||||
| TrEMBL | A0A2G5F9Q3 | 0.0 | A0A2G5F9Q3_AQUCA; Uncharacterized protein | ||||
| STRING | Aquca_001_00306.1 | 0.0 | (Aquilegia coerulea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G52150.1 | 0.0 | HD-ZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aqcoe7G157600.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



