 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aqcoe7G157600.2.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Ranunculales; Ranunculaceae; Thalictroideae; Aquilegia
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 829aa MW: 90461.9 Da PI: 6.3293 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 58.5 | 1.1e-18 | 18 | 76 | 3 | 57 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHC....TS-HHHHHHHHHHHHHHHHC CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkkl....gLterqVkvWFqNrRakekk 57
k ++t+eq+e+Le+l++++++ps +r++L +++ +++ +q+kvWFqNrR +ek+
Aqcoe7G157600.2.p 18 KYVRYTPEQVEALERLYHECPKPSSIRRQQLIRECpilsNIEPKQIKVWFQNRRCREKQ 76
5679*****************************************************97 PP
| |||||||
| 2 | START | 169.2 | 2.8e-53 | 151 | 359 | 2 | 205 |
HHHHHHHHHHHHHHHC-TT-EEEEEXCCTTEEEEEEESSS.SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-SEEEEEEEECTT..EEE CS
START 2 laeeaaqelvkkalaeepgWvkssesengdevlqkfeeskvdsgealrasgvvdmvlallveellddkeqWdetlakaetlevissg..gal 91
+aee+++e+++ka+ ++ Wv+++ +++g++++ +++ s++++g a+ra+g+v +++ v+e+l+d++ W ++++ +++++v+ +g g++
Aqcoe7G157600.2.p 151 IAEETLTEFLSKATGTAVEWVQMPGMKPGPDSIGIIAISHGCPGVAARACGLVGLEPT-RVAEILKDRPSWFRDCRVVDVMNVLPTGtgGTI 241
789*******************************************************.8888888888*********************** PP
EEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--....-TTSEE-EESSEEEEEEEECTCEEEEEEEE-EE--SSXXHH CS
START 92 qlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppe...sssvvRaellpSgiliepksnghskvtwvehvdlkgrlphw 179
+l +++l+a+++l++ Rdf+ +Ry+ l++g++v++++S++s+q p+ +++vRae+lpSg+l++p+++g+s +++v+h+dl+ +++++
Aqcoe7G157600.2.p 242 ELLYMQLYAPTTLASaRDFWLLRYTSVLEDGSLVVCERSLSSTQGGPSmplVQHFVRAEMLPSGYLVRPCEGGGSIIHIVDHMDLEPSSVPE 333
***********************************************999999*************************************** PP
HHHHHHHHHHHHHHHHHHHHTXXXXX CS
START 180 llrslvksglaegaktwvatlqrqce 205
+lr+l++s ++ ++kt++a+l+++++
Aqcoe7G157600.2.p 334 VLRPLYESPTVLAQKTTMAALRQLRQ 359
**********************9876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 1.0E-18 | 7 | 76 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 15.742 | 13 | 77 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.2E-15 | 15 | 81 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 5.13E-17 | 17 | 81 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 9.13E-17 | 18 | 78 | No hit | No description |
| Pfam | PF00046 | 2.9E-16 | 19 | 76 | IPR001356 | Homeobox domain |
| CDD | cd14686 | 1.50E-6 | 70 | 109 | No hit | No description |
| PROSITE profile | PS50848 | 24.064 | 141 | 341 | IPR002913 | START domain |
| CDD | cd08875 | 5.63E-77 | 145 | 361 | No hit | No description |
| SMART | SM00234 | 3.7E-38 | 150 | 360 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 1.19E-35 | 150 | 360 | No hit | No description |
| Gene3D | G3DSA:3.30.530.20 | 1.2E-20 | 150 | 356 | IPR023393 | START-like domain |
| Pfam | PF01852 | 7.2E-51 | 151 | 359 | IPR002913 | START domain |
| Pfam | PF08670 | 1.0E-52 | 685 | 827 | IPR013978 | MEKHLA |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0010014 | Biological Process | meristem initiation | ||||
| GO:0010075 | Biological Process | regulation of meristem growth | ||||
| GO:0010087 | Biological Process | phloem or xylem histogenesis | ||||
| GO:0048263 | Biological Process | determination of dorsal identity | ||||
| GO:0080060 | Biological Process | integument development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 829 aa Download sequence Send to blast |
MMAITSCKDG KGQMDNGKYV RYTPEQVEAL ERLYHECPKP SSIRRQQLIR ECPILSNIEP 60 KQIKVWFQNR RCREKQRKEA SRLQAVNRKL SAMNKLLMEE NDRLQKQVSQ LTAAALATTD 120 TSCESVVTSG QHHLTPQHPP RDASPAGLLS IAEETLTEFL SKATGTAVEW VQMPGMKPGP 180 DSIGIIAISH GCPGVAARAC GLVGLEPTRV AEILKDRPSW FRDCRVVDVM NVLPTGTGGT 240 IELLYMQLYA PTTLASARDF WLLRYTSVLE DGSLVVCERS LSSTQGGPSM PLVQHFVRAE 300 MLPSGYLVRP CEGGGSIIHI VDHMDLEPSS VPEVLRPLYE SPTVLAQKTT MAALRQLRQI 360 ALEASQSTVT GWGRRPAALR ALSQRLSRGF NEALNGFTDD GWSPMGSDGM DDVTILVNSS 420 PGKMSGVNIA YGNGFPAVTS GILCAKASML LQNVPPPILL RFLREHRSEW ADSNIDAYSA 480 AALKSGPCIL PGSRVGGFGG QVILPLAHTI EHEEFLEVIK LESIGHCQED TLMSRDMFLL 540 QLCSGVDENA VGTCAELVFA PIDSSFADEA PLLPSGFRII PLDFGVDASS PNRTLDLASA 600 LEIGPAGNKV PGDYSGSCGS MRSVMTIAFQ FAFESHLQEN VASMARQYLR SIISSVQRVA 660 LALSPSLLSP HSGLRPPPGT PEARTLARWI CQSYRCYLGV ELLKPTTDGS ESNLKTLWHH 720 SDAIMCCSLK ALPVFTFANQ AGLDMLETTL VALQDITLEK IFDDHGRKTL CSEFPQIMQQ 780 GFACLQGGIC LSSMGRPVSY ERAVAWKVLN EEENAHCICF MFVNWSFV* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of meristem development to promote lateral organ formation. May regulates procambial and vascular tissue formation or maintenance, and vascular development in inflorescence stems. {ECO:0000269|PubMed:15598805, ECO:0000269|PubMed:15705957, ECO:0000269|PubMed:16617092}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00200 | DAP | Transfer from AT1G52150 | Download |
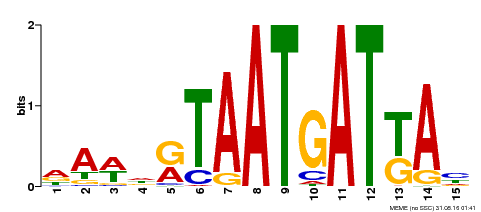
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By auxin. Repressed by miR165 and miR166. {ECO:0000269|PubMed:15773855, ECO:0000269|PubMed:16033795, ECO:0000269|PubMed:16617092, ECO:0000269|PubMed:17237362}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010267183.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein ATHB-15-like | ||||
| Refseq | XP_010267184.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein ATHB-15-like | ||||
| Swissprot | Q9ZU11 | 0.0 | ATB15_ARATH; Homeobox-leucine zipper protein ATHB-15 | ||||
| TrEMBL | A0A2G5F9R6 | 0.0 | A0A2G5F9R6_AQUCA; Uncharacterized protein | ||||
| STRING | Aquca_001_00306.1 | 0.0 | (Aquilegia coerulea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G52150.1 | 0.0 | HD-ZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aqcoe7G157600.2.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



