 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aqcoe7G263600.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Ranunculales; Ranunculaceae; Thalictroideae; Aquilegia
|
||||||||
| Family | GATA | ||||||||
| Protein Properties | Length: 311aa MW: 35004.6 Da PI: 5.33 | ||||||||
| Description | GATA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GATA | 53.3 | 3.9e-17 | 236 | 272 | 1 | 35 |
GATA 1 CsnCgttk..TplWRrgpdgnktLCnaCGlyyrkkgl 35
C++Cg ++ Tp++Rrgpdg++tLCnaCGl+++ kg+
Aqcoe7G263600.1.p 236 CRHCGISEksTPMMRRGPDGPRTLCNACGLFWANKGT 272
*****99999************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51320 | 13.69 | 95 | 130 | IPR010399 | Tify domain |
| SMART | SM00979 | 5.2E-9 | 95 | 130 | IPR010399 | Tify domain |
| Pfam | PF06200 | 2.3E-12 | 100 | 129 | IPR010399 | Tify domain |
| Pfam | PF06203 | 6.5E-17 | 163 | 205 | IPR010402 | CCT domain |
| PROSITE profile | PS51017 | 13.592 | 163 | 205 | IPR010402 | CCT domain |
| PROSITE profile | PS50114 | 10.541 | 230 | 287 | IPR000679 | Zinc finger, GATA-type |
| SMART | SM00401 | 3.5E-14 | 230 | 283 | IPR000679 | Zinc finger, GATA-type |
| SuperFamily | SSF57716 | 4.56E-13 | 232 | 285 | No hit | No description |
| CDD | cd00202 | 8.46E-15 | 235 | 280 | No hit | No description |
| Gene3D | G3DSA:3.30.50.10 | 4.6E-15 | 235 | 277 | IPR013088 | Zinc finger, NHR/GATA-type |
| Pfam | PF00320 | 3.8E-15 | 236 | 272 | IPR000679 | Zinc finger, GATA-type |
| PROSITE pattern | PS00344 | 0 | 236 | 263 | IPR000679 | Zinc finger, GATA-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 311 aa Download sequence Send to blast |
MHRVTHHTEQ ITEIRASNVG NGKNIESNYR QRGNGMMPFR NGDDMDDDEE EEVEEEEEEE 60 EEEEEEGEES EGVEGDVRLS DNGNLSDQQT LAVTDGNVGN QLTLSFQGEV YVFDSVSPEK 120 VQAVLLLLGG REIPVGVPAL PISSHHNIRE LSHVPQRLDV PQRLASLMRF REKRKERNFD 180 KKIRYSVRKE VAQRMKRHKG QFTSSKPTYE GSASVLSWDS PQWNPDGNGF QQNSTCRHCG 240 ISEKSTPMMR RGPDGPRTLC NACGLFWANK GTLRQLSKTK RMPLQNHPSS KVNKEETTAN 300 NPDAKNGLDS * |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that specifically binds 5'-GATA-3' or 5'-GAT-3' motifs within gene promoters. {ECO:0000250, ECO:0000269|PubMed:14966217}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00369 | DAP | Transfer from AT3G21175 | Download |
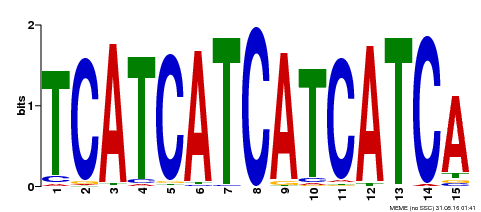
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010276694.1 | 1e-104 | PREDICTED: GATA transcription factor 28 isoform X1 | ||||
| Swissprot | Q8H1G0 | 5e-84 | GAT28_ARATH; GATA transcription factor 28 | ||||
| TrEMBL | A0A2G5DJP0 | 0.0 | A0A2G5DJP0_AQUCA; Uncharacterized protein | ||||
| STRING | Aquca_018_00034.1 | 0.0 | (Aquilegia coerulea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G21175.2 | 2e-85 | ZIM-like 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aqcoe7G263600.1.p |



