 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aqcoe7G439900.2.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; stem eudicotyledons; Ranunculales; Ranunculaceae; Thalictroideae; Aquilegia
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 387aa MW: 42871.5 Da PI: 5.0123 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 61.7 | 1.6e-19 | 110 | 159 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
+y+G+r+++ +g+W+AeIrdp++ r++lg+f taeeAa+a++a +++++g
Aqcoe7G439900.2.p 110 QYRGIRQRP-WGKWAAEIRDPRK---GVRVWLGTFNTAEEAARAYDAEARRIRG 159
69*******.**********954...4*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF54171 | 3.86E-22 | 110 | 168 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 4.8E-37 | 110 | 173 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 1.09E-32 | 110 | 168 | No hit | No description |
| PROSITE profile | PS51032 | 23.96 | 110 | 167 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 3.0E-32 | 110 | 169 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.2E-11 | 111 | 122 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 4.2E-12 | 111 | 159 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.2E-11 | 133 | 149 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0070483 | Biological Process | detection of hypoxia | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 387 aa Download sequence Send to blast |
MCGGAIISDF IPTSRSHADY LWPDLKKKKK SKKFSKPSFV PHFVDVNDDD FEADFQGFKD 60 DDSDVTEDEL VQSFSFAPKS ALPREKTTAV KSVEYNGPAE KSAKRKRKNQ YRGIRQRPWG 120 KWAAEIRDPR KGVRVWLGTF NTAEEAARAY DAEARRIRGN KAKVNFPEEN FPTAQNRTVK 180 TNSQKALPKA SQTIENPILN QDFSIMNYSS EGFNDSLGFV EEKPVIKGSG IMSRFAAMKD 240 QNGLQSFTPS DGGALCFTSD QGSNSFNCSD FGWGDHESKT PEITSVLSTT TEGNESAEYM 300 EAANPCKKLK TNSGVAAPFE GNTAKNLSEE LSAFESQMKF LQIPYLEGSW DSSMDSLFSG 360 DTTQDGGNSM DLWSFEDFPQ MVGGVF* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gcc_A | 1e-21 | 110 | 167 | 2 | 60 | ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00204 | DAP | Transfer from AT1G53910 | Download |
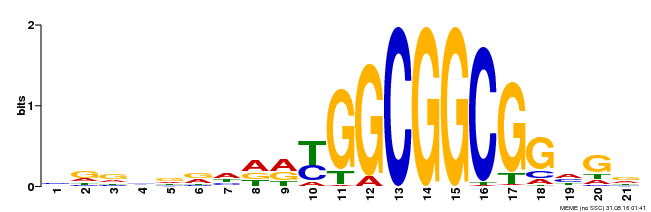
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010261706.1 | 1e-154 | PREDICTED: ethylene-responsive transcription factor RAP2-12 | ||||
| Refseq | XP_010261707.1 | 1e-154 | PREDICTED: ethylene-responsive transcription factor RAP2-12 | ||||
| TrEMBL | A0A2G5DI80 | 0.0 | A0A2G5DI80_AQUCA; Uncharacterized protein | ||||
| STRING | Aquca_020_00575.1 | 0.0 | (Aquilegia coerulea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G53910.3 | 9e-74 | related to AP2 12 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Aqcoe7G439900.2.p |



