 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aradu.1ES3P | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Dalbergieae; Arachis
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 431aa MW: 46815.8 Da PI: 6.4589 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 28.3 | 3.8e-09 | 233 | 291 | 5 | 63 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklksev 63
kr++r NR +A rs +RK +i+eLe+kv++L++e ++L +l l+ l se+
Aradu.1ES3P 233 KRAKRIWANRQSAARSKERKMRYIAELERKVQTLQTEATSLSAQLTLLQRDTNGLNSEN 291
9**********************************************999988888876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 1.7E-15 | 229 | 293 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 11.001 | 231 | 294 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 5.3E-9 | 233 | 291 | IPR004827 | Basic-leucine zipper domain |
| Gene3D | G3DSA:1.20.5.170 | 2.3E-10 | 233 | 288 | No hit | No description |
| SuperFamily | SSF57959 | 1.97E-10 | 233 | 287 | No hit | No description |
| CDD | cd14703 | 7.27E-27 | 234 | 283 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 431 aa Download sequence Send to blast |
MDKDKPLVHG GGLPPPSGRY PGYSPTGNAF NVKSEPSSSS SYPPMVPGSS PDSSHFGHGM 60 SSDSSGFSHD ISRMPDNPPR NRGHRRAHSE ILTLPDDISF DSDLGVVGGA DVPSFSDDTE 120 EDLLSMYLDM DKFNSSSATS TFQMGEPSNA AAASGSAPTS APASGGPTSS AENSVLGNNE 180 RPRVRHQHSQ SMDGSTTIKP EMLVSGSEDI SAADSKKAMS AAKLAELALI DPKRAKRIWA 240 NRQSAARSKE RKMRYIAELE RKVQTLQTEA TSLSAQLTLL QRDTNGLNSE NNELKLRLQT 300 MEQQVHLQDA LNDALKEEIQ HLKILTGQAM PPNNGGPMMN FASFGGGQQF YPNNHAMHTL 360 LAAQQFQQLQ IHPQKQQQQH QFQQLQQQML QQQQEQMHQQ SGELKIRSST PSPGPKDNAS 420 SDVNPSMGKD C |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Putative transcription factor with an activatory role. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00126 | DAP | Transfer from AT1G06070 | Download |
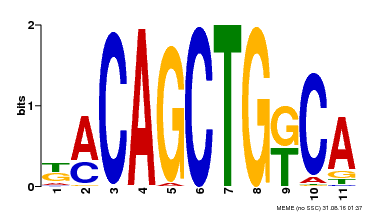
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Aradu.1ES3P |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015935496.1 | 0.0 | probable transcription factor PosF21 | ||||
| Refseq | XP_025613761.1 | 0.0 | probable transcription factor PosF21 | ||||
| Swissprot | Q04088 | 1e-155 | POF21_ARATH; Probable transcription factor PosF21 | ||||
| TrEMBL | A0A445BV43 | 0.0 | A0A445BV43_ARAHY; Uncharacterized protein | ||||
| STRING | EMJ23579 | 0.0 | (Prunus persica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF5517 | 32 | 49 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G06070.1 | 1e-141 | bZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



