 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Aradu.30PYG | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Dalbergieae; Arachis
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 861aa MW: 93608.5 Da PI: 6.3715 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 65.5 | 7.2e-21 | 153 | 208 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t++q++eLe+lF+++++p++++r eL+k+l L++rqVk+WFqNrR+++k
Aradu.30PYG 153 KKRYHRHTPQQIQELESLFKECPHPDEKQRLELSKRLCLETRQVKFWFQNRRTQMK 208
688999***********************************************999 PP
| |||||||
| 2 | START | 198.8 | 2.4e-62 | 372 | 597 | 4 | 206 |
HHHHHHHHHHHHHC-TT-EEEE.....EXCCTTEEEEEEESSS........SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S....EEEEEEEE CS
START 4 eeaaqelvkkalaeepgWvkss.....esengdevlqkfeeskv.......dsgealrasgvvdmvlallveellddkeqWdetla....kaetlevi 85
++a++elvk+a++ ep+W +s e +n +e++++f+ + +ea+r++g+v+ ++ lve+l+d + +W e+++ + +t+evi
Aradu.30PYG 372 LAAMDELVKMAQSCEPLWIRSLeiggrEVLNHEEYIRTFPNP-CialrtnsFVSEASRETGLVIINSLALVETLMDAN-RWAEMFPcmiaRTSTTEVI 467
789************************9***********664.259999999**************************.******************* PP
CTT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--...-TTSEE-EESSEEEEEEEECTCEEEEEEEE-EE--S CS
START 86 ssg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppe..sssvvRaellpSgiliepksnghskvtwvehvdlkg 174
s+g galqlm aelq+lsplvp R++ f+R+++q+ +g+w++vdvS+d ++ ++ + +v +++lpSg+++++++ng+skvtwveh+++++
Aradu.30PYG 468 SNGingtrnGALQLMHAELQVLSPLVPvREVNFLRFCKQHAEGVWAVVDVSIDAIRETSSagAPTFVNCRRLPSGCIVQDMPNGYSKVTWVEHAEYEE 565
********************************************************9999999*********************************** PP
SXXHHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 175 rlphwllrslvksglaegaktwvatlqrqcek 206
r +h+l+r l++sg+a+ga++w atlqrqce+
Aradu.30PYG 566 REVHQLYRALLSSGMAFGAQRWIATLQRQCEC 597
******************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 5.56E-21 | 135 | 210 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 4.8E-22 | 138 | 210 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.556 | 150 | 210 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 3.8E-17 | 151 | 214 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 2.78E-18 | 152 | 210 | No hit | No description |
| Pfam | PF00046 | 3.1E-18 | 153 | 208 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 185 | 208 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 44.034 | 360 | 600 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 5.68E-34 | 361 | 597 | No hit | No description |
| CDD | cd08875 | 2.04E-120 | 364 | 596 | No hit | No description |
| SMART | SM00234 | 1.1E-43 | 369 | 597 | IPR002913 | START domain |
| Pfam | PF01852 | 7.0E-54 | 372 | 597 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 2.53E-19 | 625 | 854 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009827 | Biological Process | plant-type cell wall modification | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0043481 | Biological Process | anthocyanin accumulation in tissues in response to UV light | ||||
| GO:0048765 | Biological Process | root hair cell differentiation | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 861 aa Download sequence Send to blast |
MSFGGFLDNN SGGGDDGGGG GGVRNIVEIP YNNGGGGTTK NNIINNDNNR TTMPSGAISQ 60 PRLLTTTPTL AKSMFNSPGL SLALQQTNLD GQGDVNNNNI MGSENNNNNY EGSNGGLRRS 120 REEEHESRSG SDNMDGASGD EQDAADNNNP KRKKRYHRHT PQQIQELESL FKECPHPDEK 180 QRLELSKRLC LETRQVKFWF QNRRTQMKTQ LERHENTLLR QENDKLRAEN MSIRDAMRNP 240 MCSNCGGPAI IGELSLEEQH LRIENARLKD ELDRVCALAG KFLGRPISSM GGPHPLPNSS 300 LELGVGNNNN GYGCNLSINV PNSLPLGPEF GVNLSSPLGM VSGPAITRPQ PSAVVGGFDR 360 SMERSMLLEL GLAAMDELVK MAQSCEPLWI RSLEIGGREV LNHEEYIRTF PNPCIALRTN 420 SFVSEASRET GLVIINSLAL VETLMDANRW AEMFPCMIAR TSTTEVISNG INGTRNGALQ 480 LMHAELQVLS PLVPVREVNF LRFCKQHAEG VWAVVDVSID AIRETSSAGA PTFVNCRRLP 540 SGCIVQDMPN GYSKVTWVEH AEYEEREVHQ LYRALLSSGM AFGAQRWIAT LQRQCECLAI 600 LMSSTQPSRD HSAITASGRR SMLKLAQRMT NNFCAGVCAS TVHKWNKLNN TGNTXXXXXX 660 XXXXXXXIVL SAATSVWLPV SPQRLFDFLR DERLRSEWDI LSNGGPMQEM AHIAKGQDHG 720 NCVSLLRAGA INSNQSSMLI LQETCIDAAG SLVVYAPVDI PAMHVVMNGG DSAYVALLPS 780 GFAIVPDGPA PRGAQNGGAD ANGGDDLGGG ARGSGSLLTV AFQILVNSLP TAKLTVESVE 840 TVNNLISCTV QKIKAALRCE S |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of the tissue-specific accumulation of anthocyanins and in cellular organization of the primary root. {ECO:0000269|PubMed:10402424}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00421 | DAP | Transfer from AT4G00730 | Download |
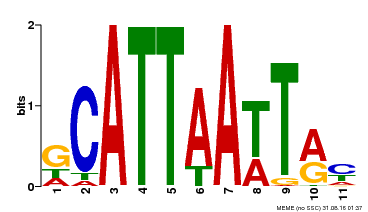
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Aradu.30PYG |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020991942.1 | 0.0 | homeobox-leucine zipper protein ANTHOCYANINLESS 2 | ||||
| Swissprot | Q0WV12 | 0.0 | ANL2_ARATH; Homeobox-leucine zipper protein ANTHOCYANINLESS 2 | ||||
| TrEMBL | A0A445EGZ0 | 0.0 | A0A445EGZ0_ARAHY; Uncharacterized protein | ||||
| STRING | XP_007134961.1 | 0.0 | (Phaseolus vulgaris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1192 | 34 | 106 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00730.1 | 0.0 | HD-ZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



